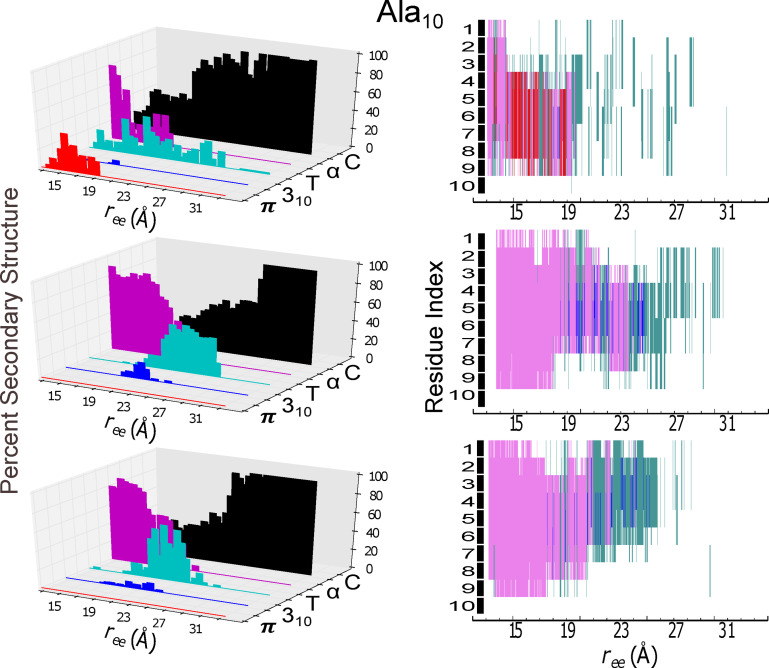
Figure 4.
Secondary structure content (panels at left) and individual residue structural assignment (panels at right) for the model helical peptide are shown as a function of extension, . The trajectory for the protein in explicit solvent obtained at 1 Å/ns with ASMD that was nearest to the computed Jarzynski average was chosen for analysis using the STRIDE algorithm in VMD in c22 (top panels), in c27 (middle panels), and in c36 (bottom panels). The evolution of the secondary structure character as a function of mechanical unfolding is displayed as: π‐helical (red), 310‐helical (blue), turn (T in teal), α‐helical (purple), and random coil (C in black). In the right column panels, the secondary structural assignments of each amino acid residue is indexed from 1 to 10, starting with the amino‐terminal (N) end.