Background:
The purpose of this study was to evaluate existing evidence in the field of long non-coding RNAs (lncRNAs) and prognosis of gastric cancer.
Methods:
A comprehensive literature search was performed through the electronic database. The combined hazard ratios (HRs) and corresponding 95% confidence intervals (CIs) of overall survival (OS), disease-free survival (DFS), or progression free survival (PFS) were calculated to assess the strength of the association. Kaplan–Meier (KM) plotter was used to verify lncRNA HOX transcript antisense RNA (HOTAIR) expression and OS.
Results:
Overall, a significant correlation between high lncRNAs expression and poor OS was explored in patients with gastric cancer (HR = 1.78, P < .001). Subgroup analysis based on statistical methods indicated the high expression of lncRNAs in log-rank (HR = 1.87, P < .001) and multivariate analysis (HR = 1.71, P < .001) were all significantly correlated with the poor OS. Clinicopathological parameters analysis showed the lncRNA expression were significantly associated prognosis, including TNM stage, tumor size, pathological differentiation, lymph nodes metastasis, distance metastasis, invasion depth and Lauren’s classification. It was consistent with the verification results of bioinformatics database for lncRNA HOTAIR (P < .001).
Conclusion:
Our study confirmed the expression of lncRNAs and clinicopathological features may serve as effective indicators of prognosis in patients with gastric cancer.
Keywords: bioinformatics, clinicopathological factors, field synopsis, gastric cancer, lncRNAs, prognosis
1. Introduction
Gastric cancer is a highly lethal malignancy tumor in the world. It is the fourth most common cancer and the second leading cause of cancer death.[1,2] The incidence rate and mortality rate in East Asia is the highest in the worldwide.[3] Although surgical operation, chemotherapy and radiotherapy regimens can help reduce the incidence and mortality rates of gastric cancer, the overall 5 year survival rate is still only about 25%.[4,5]
In recent years, with the extensive research of gastric cancer coding proteins, the research of long noncoding RNA (lncRNAs) has attracted more and more attention.[6] The lncRNAs are transcribed RNA with a length of more than 200 nt and cannot encode proteins.[7] LncRNAs have attracted attention in different studies on transcriptional, post-transcriptional, and epigenetic levels to modify the expression of protein coding genes,[8] and they regulate diverse biological processes, such as signal transduction, cellular functions, proliferation, differentiation, and apoptosis.[9–11]
Emerging evidence has demonstrated the significance of dysregulated lncRNAs expression in malignancies[12] and lncRNAs as oncogenes or tumor suppressors in pathological processes of cancer.[13,14] Recent studies have evaluated the abnormal lncRNAs expression in gastric cancer and test in clinical correlated diagnosis, treatment and prognostic prediction.[6,14–17] It was believed that specific changes of lncRNAs could make them ideal biomarkers for prognosis of gastric cancer.[18] Therefore, the clinical value of lncRNAs need to identify and to predict the prognosis in gastric cancer patients.
The role of lncRNAs in the prevention of gastric cancer remains elusive. A comprehensive meta-analysis of clinical evidence will help resolve the critical issue on whether lncRNAs could predict the prognosis of gastric cancer. So far, there was one related systematic review[19] published in 2017, which reported a similar question. It only included 38 studies ranging from 2014 to 2016.
The Meta-analysis of gastric cancer-related lncRNA has mainly focused on the systematic evaluation of single lncRNA[20,21] and the risk of gastric cancer or the prognosis of lncRNA and different cancers.[21,22] In addition to including more recent studies, in our study, a 2-sample Z-test was used to see whether the HR differ by log rank or multivariate analysis, Meanwhile, we compared the results with log rank and multivariate analysis to verify the reliability and stability of the results. For the clinicopathological parameters, the main evaluating indicator is different. We pooled all the clinicopathological data from the multivariate analysis results of the included studies, the evaluating indicator was HR. But Zhu M, et al study used OR to explore the correlation between lncRNAs transcription level and clinicopathological parameters, it ignores the influence of survival time factors.
The aim of our work is to fill the gap in the world published medical literature by performing the systematic synopsis of the available evidence in the sphere of lncRNAs and prognosis of gastric cancer including the overall survival (OS), disease-free survival (DFS), progression-free survival (PFS), Disease Specific Survival (DSS), recurrence-free survival (RFS) or clinicopathological factors. Furthermore, we used Kaplan–Meier (KM) plotter to verify the effect of lncRNA HOTAIR expression on survival using 631 gastric cancer patients with OS.
2. Materials and Methods
The present study was performed according to the guidelines of the Meta-analysis of Observational Studies in Epidemiology.[23] In the process of constructing the clinical significance of gastric cancer related lncRNA, we completed the study design based on the principles of population, intervention, comparison, outcome, and research design.
2.1. Search strategy
A 2-step search strategy was adopted. The first step a systematic review of original articles was performed by searching PubMed, Excerpta Medica database, Web of Science, and Chinese database (Wanfang and China national knowledge infrastructure) to April 16, 2021. The search included the following 3 groups of terms: “tumor,” and “cancer” “carcinoma”; “stomach” and “gastric”; “lncRNA,” ‘‘long noncoding RNA”; ‘‘lincRNA,” ‘‘long intergenic non-coding RNA”; “prognosis” and “survival.” Searches were conducted using all combinations of at least one term from each group.
The second step, we manually retrieved bibliography of relevant articles to further determine the potential studies that has not been retrieved by databases exploration. Two investigators (FJD and XNX) performed this comprehensive online search.
2.2. Inclusion and exclusion criteria
The included studies met the following criteria: cohort studies that investigated associations between lncRNAs expression and gastric cancer with OS, DFS, RFS, PFS, and/or clinicopathological features, gastric cancer were divided into 2 groups according to the high and low expression of lncRNAs, hazard ratios (HRs) and 95%CIs were reported or could be calculated from given data, published in English or Chinese. Exclusion criteria for the articles included: letters, reviews, expert opinions, meta-analysis and case reports, duplicate publications; studies with insufficient data to calculate the HRs and 95%CIs.
If the data overlaps with other published literature, we select a newly published and /or larger sample article.
2.3. Data extraction
The data included in the study were independently evaluated by 2 authors (FJD and XNX). If there were different opinions, it should be determined after consultation with the third author (KJW).
The data of the following items were extracted: The name of first author’s, year of publication, ages and genders, sample size, follow-up time, pathology subtypes, clinicopathological characteristics, OS and DFS/ PFS/DSS/RFS and 95%CIs. For the OS, the starting point is diagnostic time, and operation day or treatment time for others. When HR and/or 95%CI were not reported, the methods of Parmar[24] and Tierney[25] were used for extrapolation.
2.4. LncRNA HOTAIR expression profile and prognosis
The KM plotter is a summary analysis based on biomarker evaluation to measure the effect of lncRNA HOTAIR expression level on OS in 631 gastric patients.[26] Meanwhile, the statistical correlation and the visualization of cutoff value were presented.
2.5. Statistical analysis
The pooled HRs with 95%CIs were conducted by Revman 5.3.5 (Cochrane Collaboration, Oxford, UK) to evaluate the relationship between lncRNA expression, clinicopathological features and prognosis. Inter-study heterogeneity was quantified using Q tests and the I-squared (I2) statistic.[27] According to the results of heterogeneity analysis, a fix effects or random model was performed. In the absence of significant heterogeneity (Pheterogeneity ≥ 0.10 or I2 ≤ 50%), a fixed-effects model[28] was applied to assess the combined effect size, otherwise (Pheterogeneity < 0.1 and I2 > 50%) the random-effects model[29] was conducted. For studies that did not report the HRs with 95% CIs, Engauge Digitizer 10.0 (https://sourceforge.net/projects/digitizer/) was used to deduce the original data from a KM curve. Subgroup analyses were performed by different prognoses, regions, cutoff values and clinicopathological features. Begg’s[30] and Egger’s test[31] were used to assess the publication bias in STATA 13.1MP.
All P values were 2-sided and P < .05 was considered statistically significant. The KM plotter split is median, and the lncRNA HOTAIR expression profile from gastric cancer samples and paired normal tissues.
3. Results
3.1. Characteristics of eligible studies
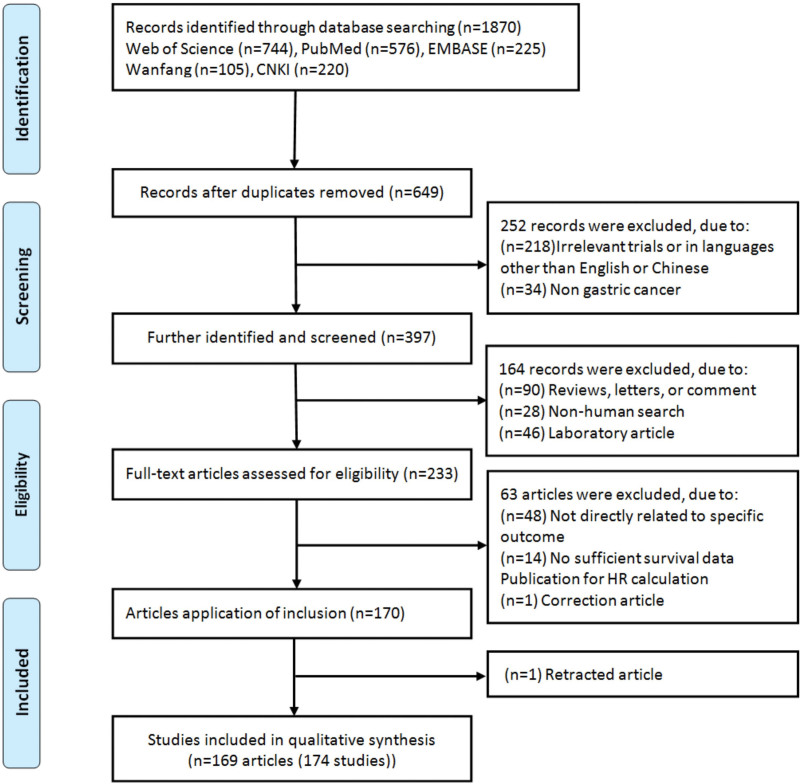
A total of 1870 records were retrieved according to the literature search strategy. According to the inclusion and exclusion criteria, 1 article[32] was eliminated due to significant overlap with a previously published article by Xu et al.[33] As a result, we identified 169 eligible articles (174 studies) (Table S1, Supplemental Digital Content, http://links.lww.com/MD/H400) comprising 16280 patients (range of sample size: 32–373, mean: 100) with gastric cancer. Three articles[34–36] performed 2 cohort studies in different populations, 1 article[37,38] included 2 pathological types of gastric cancer, and we considered them as 2 studies, respectively. The details on the flow diagram of the literature search strategy was presented in Figure 1. Three articles[35,39,40] contained 2 lncRNAs, and the other 2 articles[36,41] contain 12 and 5 lncRNAs, respectively (Table S1, Supplemental Digital Content, http://links.lww.com/MD/H400). Therefore, a total of 185 gastric cancer related lncRNAs were analyzed. All included studies were from 2013 to 2021. Based on the study of regional sources, 97.1% of the studies were Chinese (169/174). Four eligible studies were specified the histological subtypes (intestinal, diffuse and adenocarcinoma) of gastric cancer (n = 5, 2.9%). The methods of lncRNA detection in tissue (n = 169, 97.1%) or serum (n = 5, 2.9%) were quantitative real-time polymerase chain reaction (qRT-PCR). The cutoff values of lncRNA were most taken as the median (n = 102, 58.6%) (Table S1, Supplemental Digital Content, http://links.lww.com/MD/H400).
Figure 1.
Flow chart of literature search and study selection.
3.2. Analysis of deviation statistics
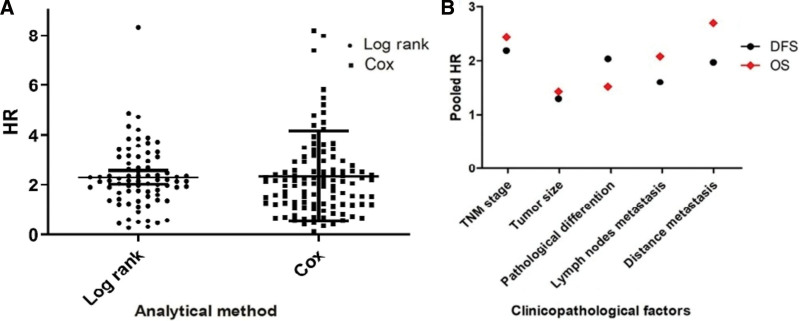
A 2-sample Z test was used to detected whether the HR differ by log rank (KM Curve) or multivariate analysis (Cox-analysis), there was no statistically significant difference between the 2 groups (Z = 0.2336,P = .8155, Fig. 2A). T test was used to identified difference between clinicopathological features of OS and DFS, and the results showed no statistical significance (P = .3625, Fig. 2B).
Figure 2.
(A) Z test between the Log rank and multivariate analysis. (B) T test for the OS and DFS of clinicopathological features. DFS = disease-free survival, OS = overall survival.
3.3. Association between lncRNA expression level and OS
One hundred and seventy-four studies (185 lncRNAs dataset) were included for the analysis of correlation between expression of lncRNA and OS in patients with gastric cancer. Obvious heterogeneity existed (I2 = 87%, Pheterogeneity < 0.001) among 165 studies (185 lncRNAs dataset). Therefore, a random effect was applied, and a significant correlation between high lncRNAs expression and poor OS in patients with gastric cancer was explored (HR = 1.78, 95% CI: 1.64–1.93, P < .001). Subgroup analysis was performed according to statistical methods, the results showed that the high lncRNAs expression were significantly correlated with poor OS in log rank (HR = 1.87, 95%CI: 1.66–2.10, P < .001) and multivariate analysis (HR = 1.71, 95%CI: 1.55–1.89, P<.001). For subgroups analyses, the results indicated that there was no heterogeneity between subgroups (I2 = 23.6%, P = .25) (Table 1).
Table 1.
Main results of pooled HRs in the meta-analysis.
| Comparisons | Heterogeneity test | Summary HR (95% CI) |
Hypothesis test | Studies | LncRNAs Number |
|||
|---|---|---|---|---|---|---|---|---|
| I2(%) | P | Model | Z | P | ||||
| OS (Low vs High) | ||||||||
| Total | 87 | <.001 | Random | 1.78(1.64, 1.93) | 13.84 | <.001 | 165 | 185 |
| Log rank (KM) | 80 | <.001 | Random | 1.87(1.66,2.10) | 10.29 | <.001 | 75 | 78 |
| Multivariate analysis (Cox) | 85 | <.001 | Random | 1.71(1.55,1.89) | 10.53 | <.001 | 90 | 107 |
| Subgroup differences | 23.6 | .25 | ||||||
| cutoff (High vs Low) | ||||||||
| Median | 86 | <.001 | Random | 1.87(1.69,2.08) | 11.66 | <.001 | 98 | 104 |
| Normal | 84 | <.001 | Random | 1.64(1.45,1.88) | 6.47 | <.001 | 62 | 80 |
| Mean | 4.77(2.98,7.65) | 6.49 | <.001 | 1 | 1 | |||
| Country | ||||||||
| China | 87 | <.001 | Random | 1.78(1.64,1.94) | 13.67 | <.001 | 161 | 179 |
| Others | 53 | .06 | Random | 1.72(1.16,2.54) | 2.71 | .007 | 4 | 6 |
| LncRNAs | ||||||||
| LncRNA HOTAIR | 46 | <.001 | Fixed | 1.83(1.55,2.15) | 7.27 | <.001 | 10 | 11 |
| LncRNA AFAP1-AS1 | 29 | <.001 | Fixed | 2.73(1.76,4.22) | 4.50 | <.001 | 3 | 3 |
| Others | 87 | <.001 | Random | 1.75(1.61,1.90) | 12.91 | <.001 | 152 | 171 |
| Subgroup differences | 32 | .23 | ||||||
| DFS/PFS/DSS/RFS | ||||||||
| Total | 84 | <.001 | Random | 1.74(1.47,2.06) | 4.32 | <.001 | 58* | 61 |
| DFS | 80 | <.001 | Random | 1.65(1.38,1.97) | 5.56 | <.001 | 39 | 41 |
| PFS | 90 | <.001 | Random | 2.08(1.28,3.38) | 2.95 | .003 | 14 | 15 |
| DSS | 79 | .003 | Random | 1.30(0.70,2.43) | 0.83 | .40 | 4 | 4 |
| RFS | 2.76(1.42,5.36) | 2.99 | .03 | 1 | 1 | |||
| Subgroup differences | 14.4 | .32 | ||||||
Cox = survival data from a Cox-analysis, DFS = disease-free survival, DSS = disease specific survival, KM = survival data from a Kaplan–Meier curve, OS = overall survival, PFS = progressive free survival, RFS = recurrence-free survival.
Analyses of subgroups were conducted by cutoff, and the results revealed a significant correlation between high expression of lncRNAs and poor OS in median (HR = 1.87, 95%CI: 1.69–2.08, P < .001), normal (HR = 1.64, 95%CI: 1.45–1.88, P = .006) and mean subgroups (HR = 4.77, 95%CI: 2.98–7.65, P < .001). Meanwhile, our analysis revealed a positive link between high lncRNAs expression and poor OS in China (HR = 1.78, 95% CI: 1.64–1.94, P < .001) and other countries (HR = 1.72, 95% CI: 1.16–2.54, P = .007) (Table 1).
3.4. Association between lncRNA expression level and DFS/ PFS/DSS/RFS
A total of 58 studies (61 lncRNAs) were included to analyze the correlation between lncRNA expression and DFS/ PFS/DSS/RFS. There was obvious heterogeneity among studies (I2 = 84%, Pheterogeneit < 0.001), and a random-effects model was applied to calculate the pooled HR and 95%Cl. The results showed that the pooled HR of disease progression lncRNAs expression and DFS/PFS/DSS/RFS in patients with gastric cancer was 1.74 (95%CI: 1.47–2.06). Meanwhile, stratified analysis showed that the HR of the high lncRNAs expression group versus the low lncRNAs expression group in DFS, PFS, DSS and RFS were 1.65 (95%CI: 1.38–1.97), 2.08 (95%CI: 1.28–2.38), 1.30 (95%CI:0.70–2.43), and 2.76 (95%CI:1.42–5.36), respectively. For subgroups analyses, the results showed that there was no heterogeneity among these groups (I2 = 14.4%, P = .32) (Table 1).
3.5. Association between lncRNA expression and clinicopathological parameters
We combined all the clinicopathological data from the multivariate analysis results to explore the association between lncRNA expression and clinicopathological features (Table 2).
Table 2.
Multivariate analysis of independent prognostic factors of pooled HRs in the meta-analysis.
| Characteristics | Comparisons | Case number | Heterogeneity test |
Summary HR (95% CI) |
Hypothesis test | Model | Studies | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Q | P | I2(%) | Z | P | ||||||
| OS | ||||||||||
| TNM stage | III + IV vs I + II | 9014 | 233.24 | <.001 | 71 | 2.07(1.83,2.34) | 11.5 | <.001 | Random | 68 |
| Tumor size | ≥5 vs < 5 | 3390 | 38.77 | .05 | 33 | 1.32(1.21,1.45) | 5.93 | <.001 | Fixed | 27 |
| Pathological differentiation | Poor vs Well-moderate | 4429 | 65.68 | <.001 | 53 | 1.35(1.16,1.57) | 3.96 | <.001 | Random | 32 |
| Lymph nodes metastasis | Positive vs Negative | 6802 | 242.68 | <.001 | 79 | 1.90(1.58,2.28) | 6.92 | <.001 | Random | 52 |
| Distance metastasis | Positive vs Negative | 5386 | 95.85 | <.001 | 61 | 2.81(2.29,3.45) | 9.88 | <.001 | Random | 38 |
| Invasion depth | T3 + T4 vs T1 + T2 | 4153 | 97.44 | <.001 | 65 | 1.53(1.33,177) | 5.86 | <.001 | Random | 35 |
| Lauren’s classification | Diffuse vs Intestinal | 1181 | 6.43 | .49 | 0 | 1.31(1.08,1.60) | 2.73 | .006 | Fixed | 8 |
| DFS | ||||||||||
| TNM stage | III + IV vs I + II | 3264 | 17.08 | .81 | 0 | 2.32(2.02,2.66) | 12.02 | <.001 | Fixed | 24 |
| Tumor size | ≥5 vs < 5 | 932 | 3.68 | .05 | 0 | 1.05(1.00,1.10) | 1.81 | .07 | Fixed | 6 |
| Pathological differentiation | Poor vs Well-moderate | 918 | 21.84 | <.001 | 77 | 1.55(1.02,2.36 | 2.06 | .04 | Random | 6 |
| Lymph nodes metastasis | Positive vs Negative | 2066 | 19.07 | .09 | 37 | 1.81(1.56,2.10) | 7.75 | <.001 | Fixed | 13 |
| Distance metastasis | Positive vs Negative | 1829 | 5.18 | .92 | 0 | 2.20 (1.82,2.66) | 8.11 | <.001 | Fixed | 12 |
| Invasion depth | T3 + T4 vs T1 + T2 | 1389 | 29.88 | <.001 | 67 | 1.40(0.98,2.01) | 1.84 | .07 | Random | 11 |
DFS = disease-free survival, OS = overall survival.
For the OS, the results revealed that the expression of lncRNA were significantly correlated with the clinicopathological parameters, including TNM (HR = 2.07, 95%CI: 1.83–2.34, P < .001), tumor size (HR = 1.32, 95%CI: 1.21–1.45, P < .001), pathological differentiation (HR = 1.35, 95%CI: 1.16–1.57, P < .001), lymph nodes metastasis (HR = 1.90, 95%CI: 1.58–2.28, P < .001), distance metastasis (HR = 2.81, 95%CI: 2.29–3.45, P < .001),invasion depth (HR = 1.53, 95%CI: 1.08–1.60, P < .001), Lauren’s classification (HR = 1.31, 95%CI: 1.08–1.60, P = .006).
For the DFS, the results indicated that there was significantly associated with TNM stage (HR = 2.32, 95%CI: 2.02–2.66, P < .001), lymph nodes metastasis (HR = 1.81, 95%CI: 1.56-2.10, P < .001), distance metastasis (HR = 2.20, 95%CI: 1.82–2.66, P < .001), and invasion depth (HR = 1.45, 95%CI: 1.07–1.97, P = .02). However, the association of lncRNA expression with tumor size (HR = 1.05, 95%CI: 1.00–1.10, P = .07), pathological differentiation (HR = 1.55, 95%CI: 1.02–2.36, P = .04) and invasion depth (HR = 1.40, 95%CI: 0.98–2.01, P = .07) and the risk of gastric cancer was at a statistically significant threshold.
3.6. Sensitivity analyses and assessment of publication bias
Sensitivity analysis was implemented by omitting removing a study at one time, and recalculating the combined HR, the pooled HR was not substantially changed, which indicated the combined HR was stable (Data not shown).
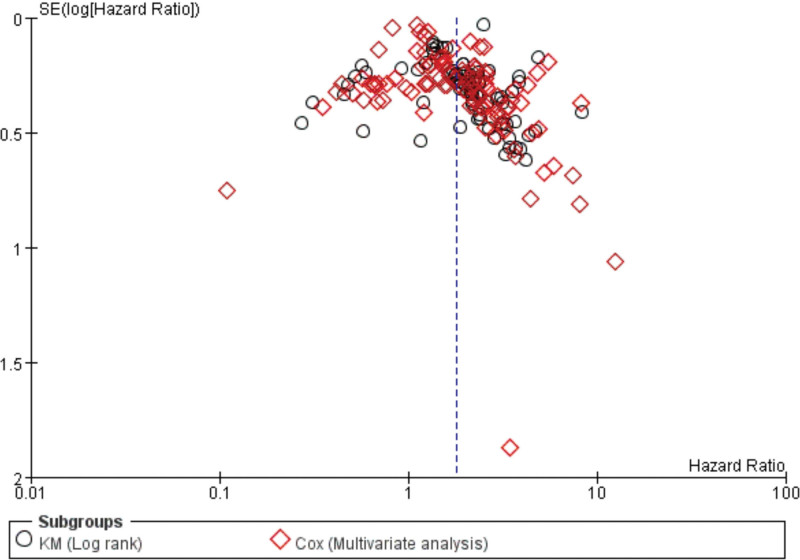
Begg’s and Egger’s tests were used to estimate the bias of publication. The results didn’t explore any evidence of publication bias (Table 3), and the funnel plots shape was basically symmetrical (Fig. 3).
Table 3.
Publication bias of lncRNA for Begg’s test and Egger’s test.
| Comparisons | Begg’s test | Egger’s test | |||
|---|---|---|---|---|---|
| z | P | t | P | 95% CI | |
| OS-Combine | 2.50 | .013 | 2.55 | .012 | 0.257-2.063 |
| Log rank (KM) | 1.67 | .097 | -0.81 | .075 | -1.411-0.681 |
| Multivariate analysis (Cox) | 1.73 | .084 | 1.76 | .083 | -0.121-1.912 |
| PFS | 1.64 | .101 | 1.55 | .132 | -0.588-0.958 |
| DFS | 0.62 | .533 | -1.29 | .229 | -6.080-4.125 |
Cox = survival data from a Cox-analysis, KM = survival data from a Kaplan–Meier curve.
Figure 3.
Begg’s funnel plot of publication bias on the relationship between lncRNA expression and OS. lncRNAs = long non-coding RNAs, OS = overall survival.
3.7. Expression of lncRNA HOTAIR and prognosis in database test
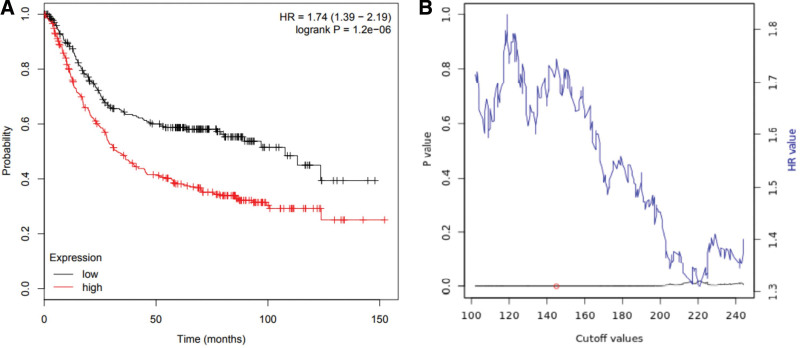
For the OS, there was a highly significant association between high expression and pool OS in gastric cancer patients (HR = 1.74, 95% CI: 1.39–2.19, P < .001) (Fig. 4A). The results of direct sequencing and lncRNA HOTAIR expression were consistent with the combined results of our individual studies. The linear relationship between statistical correlation and cutoff value was shown in Figure 4B.
Figure 4.
(A) Kaplan–Meier survival curves for OS according to lncRNA HOTAIR expression in gastric cancer patients. OS of patients with high versus low lncRNA HOTAIR expression was shown. (B) Significance versus cutoff values between lower and upper quartiles of expression. HOTAIR = HOX transcript antisense RNA, lncRNAs = long non-coding RNAs, OS = overall survival.
4. Discussion
Gastric cancer is one of the most aggressive malignant tumors,[42] and most patients with gastric cancer are diagnosed as advanced and have a poor prognosis.[43,44] In recent years, more and more evidence shows that the abnormal lncRNAs expression is related to the clinical prognosis of cancer patients. The lncRNAs have made great contributions to the mechanism, function and translation of cancer biology, and play an important role in the occurrence and progression of gastric cancer.[45] Thus, more sensitive gastric cancer biomarkers for improving screening, diagnosis and prognostic assessment are urgently needed. In order to clarify the expression significance of lncRNAs and the value of clinical pathological parameters in gastric cancer, we conducted the present field synopsis of observational studies, and then databases validation.
In order to identify lncRNAs with potential biological functions, we used log-rank and multivariate cox regression analysis to analyze the correlation between lncRNAs expression, clinicopathological characteristics, and patient overall survival. We validated that no statistically significant difference between the log-rank and multivariate analysis (P = .25). Meanwhile, the T-test was used to identified distributed difference between the OS and DFS of clinicopathological features, and the results showed no significant difference (P = .3625). The pooled analysis results showed that a significant correlation was revealed between high lncRNAs and poor OS in gastric cancer patients (HR = 1.78, 95% CI: 1.64–1.93, P < .001). In subgroup analysis, we explored that the high lncRNAs expression in both log-rank (HR = 1.87, 95%CI: 1.66–2.10, P < .001) and multivariate cox regression analysis (HR = 1.71, 95%CI: 1.55–1.89, P < .001) was significantly related with poor OS. The stratified analysis was performed by cutoff, and a positive link between elevated lncRNAs and poor OS in median, normal and mean subgroups were revealed. Meanwhile, our analysis revealed a significant correlation between high lncRNAs expression and poor OS in China and other countries.
For the DFS/ PFS/DSS/RFS, combined HR of correlation between elevated expression of lncRNAs and poor DFS/PFS/DSS/RFS in patients with gastric cancer was 1.74 (95%CI: 1.47–2.06), stratified analysis shows that association was statistically significant difference in DFS, PFS, DSS, and RFS, respectively.
In analysis of clinicopathological features and prognosis, we found significant changes in the correlation between dysregulated lncRNAs and clinicopathological characteristics. The pooled results showed that lncRNAs were significantly related to TNM stage, tumor size, pathological differention, lymph nodes metastasis, distance metastasis, invasion depth in OS, and TNM stage, lymph nodes metastasis, distance metastasis, invasion depth in DFS, but no association was explored between lncRNA expression and other clinicopathological features (Lauren’s classification in OS; tumor size and pathological differentiation in DFS). The aberrant lncRNAs expression supported the corresponding clinical value in identifying clinicopathological features, especially TNM stage.
To confirm the reliability of the results, we selected 11 studies on lncRNA HOTAIR to explore the prognostic value of lncRNA HOTAIR in GEO, EGA and TCGA, and then verify our results. We used 631 gastric cancer tissues and normal tissues in the public database. The results showed that the lncRNA HOTAIR expression was significantly higher than that in the normal control group. These findings further validate our conclusions and suggest that lncRNA HOTAIR can be used as an independent prognostic factor for OS in patients with gastric cancer.
In recent decades, accumulating evidences have shown that lncRNAs are crucial in tumorigenesis.[44] It is a functional end product, and the lncRNA expression level is directly related to the level of active molecules.[46] Using lncRNA to diagnose and assess the prognosis has intrinsic advantages compared with other protein-coding RNAs. According to the data set of the included studies in this study, 97.6% of lncRNA expression was measured in tissue. Compared to miRNAs and protein-coding mRNAs, lncRNAs show greater tissue specificity, which makes them suitable for the novel diagnostic and prognostic cancer biomarkers.[47]
Accumulating evidence indicates that lncRNAs have a biological role in regulating the occurrence and development of gastric cancer.[48–50] Furthermore, tumor specific lncRNAs may also be link to metastasis and invasion of gastric cancer. Lymph nodes metastasis, which is the most common metastasis pathway of gastric cancer, and it is of great significance for the diagnosis and prognosis, TNM staging and treatment of patients with gastric cancer. According to our results, we demonstrated that there was a significant correlation between lncRNAs expression and lymph nodes metastasis in OS and DFS, indicating that lncRNAs should be a potential bio-marker for judging lymph nodes metastasis in patients of gastric cancer.
Nevertheless, certain limitations of the present study should be presented. Firstly, most of included articles were from China, and studies from other countries might attain different outcomes. Secondly, the confounding factors induced by diverse RNA extraction methods and RNA detection platforms may limit the validity of this study. Thirdly, the diverse sample sources resulted in significant heterogeneity between individual studies, although the random-effects model was performed to reduce the influence of heterogeneity on our results. Fourthly, although no evidence of publication bias was explored, the included studies were all published in public databases, there was no unpublished data in the database we used, which may generate publication bias. Finally, in the included studies, the cutoff value of lncRNAs expression was different, and the real value may be deviated due to different algorithms.
5. Conclusions
The high expression of lncRNA can predict the poor clinical prognosis of gastric cancer, and the abnormality of lncRNA is related to the clinicopathological features, suggesting lncRNA may serve as a novel valuable biomarker for the prognostic of gastric cancer, especially lncRNA HOTAIR. Future studies need to use multiply reliable and sensitive detection methods in large-scale multicenter studies to validate the true prognostic significance of lncRNAs in patients with gastric cancer.
Author contributions
Conceptualization: Xiaona Xu.
Data curation: Fujiao Duan.
Formal analysis: Haili Wang.
Investigation: Shiutin Ng.
Project administration: Kaijuan Wang.
Software: Yilin Li.
Supervision: Erping Xu.
Visualization: Fujiao Duan.
Writing – original draft: Xiaona Xu.
Writing – review & editing: Guanghui Niu.
Supplementary Material
Abbreviations:
- CIs =
- confidence intervals
- DFS =
- disease-free survival
- DSS =
- disease specific survival
- HOTAIR =
- HOX transcript antisense RNA
- HRs =
- hazard ratios
- KM =
- Kaplan Meier
- lncRNAs =
- long non-coding RNAs
- OS =
- overall survival
- PFS =
- progression free survival
- RFS =
- recurrence-free survival
- TNM =
- tumor-node-metastasis
The datasets generated during and/or analyzed during the current study are publicly available.
This study was reported by the Preferred Reporting Items for Systematic reviews and Meta-Analyses. It was based on previous publications and therefore did not require ethical approval or informed consent.
This work was supported by grants from the National Natural Science Foundation of China (No. 81672917, 14207450), Joint Construction Project of Medical Science and Technology Program (LHGJ20210184) Henan Province young and middle-aged health science and technology innovation excellent young talent training project (YXKC2022044).
Supplemental Digital Content is available for this article.
The authors have no conflicts of interest to disclose.
How to cite this article: Xu X, Duan F, Ng S, Wang H, Wang K, Li Y, Niu G, Xu E. Clinicopathological and prognostic value of lncRNAs expression in gastric cancer: A field synopsis of observational studies and databases validation. Medicine 2022;101:40(e30817).
Contributor Information
Xiaona Xu, Email: xuerping@hactcm.edu.cn.
Fujiao Duan, Email: fjduan@126.com.
Shiutin Ng, Email: dr.ng@live.ca.
Haili Wang, Email: kjwang@163.com.
Kaijuan Wang, Email: kjwang@163.com.
Yilin Li, Email: beckylixinrui@gmail.com.
Guanghui Niu, Email: lonew@foxmail.com.
References
- [1].L SR, D MK, Ahmedin J. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30.29313949 [Google Scholar]
- [2].Torre LA, Siegel RL, Ward EM, et al. Global cancer incidence and mortality rates and trends—an update. Cancer Epidemiol Biomarkers Prev. 2016;25:16–27. [DOI] [PubMed] [Google Scholar]
- [3].Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–86. [DOI] [PubMed] [Google Scholar]
- [4].Kitayama J, Ishigami H, Yamaguchi H, et al. Treatment of patients with peritoneal metastases from gastric cancer. Ann Gastroenterol Surg. 2018;2:116–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Zhao J, Liu Y, Zhang W, et al. Long non-coding RNA Linc00152 is involved in cell cycle arrest, apoptosis, epithelial to mesenchymal transition, cell migration and invasion in gastric cancer. Cell Cycle. 2015;14:3112–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ren W, Zhang J, Li W, et al. A tumor-specific prognostic long non-coding RNA signature in gastric cancer. Med Sci Monitor Int Med J Experimental Clin Res. 2016;22:3647–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Jiang C, Xin L, Hui Z, et al. Long non-coding RNAs: potential new biomarkers for predicting tumor invasion and metastasis. Mol Cancer. 2016;15:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–9. [DOI] [PubMed] [Google Scholar]
- [9].Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. [DOI] [PubMed] [Google Scholar]
- [10].Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Strobel EJ, Watters KE, Loughrey D, et al. RNA systems biology: uniting functional discoveries and structural tools to understand global roles of RNAs. Curr Opin Biotechnol. 2016;39:182–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–61. [DOI] [PubMed] [Google Scholar]
- [13].Wagner LA, Christensen CJ, Dunn DM, et al. EGO, a novel, noncoding RNA gene, regulates eosinophil granule protein transcript expression. Blood. 2007;109:5191–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Zhang H, Chen Z, Wang X, et al. Long non-coding RNA: a new player in cancer. J Hematol Oncol 6,1(2013-05-31). 2013;6:37–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zhou X, Yin C, Dang Y, et al. Identification of the long non-coding RNA H19 in plasma as a novel biomarker for diagnosis of gastric cancer. Sci Rep. 2015;5:11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Baratieh Z, Khalaj Z, Honardoost MA, et al. Aberrant expression of PlncRNA-1 and TUG1: potential biomarkers for gastric cancer diagnosis and clinically monitoring cancer progression. Biomarkers Med. 2017;11:1077. [DOI] [PubMed] [Google Scholar]
- [17].Hu Y, Wang J, Qian J, et al. Long noncoding RNA GAPLINC regulates CD44-dependent cell invasiveness and associates with poor prognosis of gastric cancer. Cancer Res. 2015;13:e100–1. [DOI] [PubMed] [Google Scholar]
- [18].Cabanski CR, White NM, Dang HX, et al. Pan-cancer transcriptome analysis reveals long noncoding RNAs with conserved function. RNA Biol. 2015;12:628–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zhu M, Wang Y, Liu X, et al. LncRNAs act as prognostic biomarkers in gastric cancer: a systematic review and meta-analysis. Front Laboratory Med. 2017;1:59–68. [Google Scholar]
- [20].Chen W, Li Y, Guo L, et al. Long non-coding RNA FTX predicts a poor prognosis of human cancers: a meta-analysis. Biosci Rep. 2021;41:BSR20203995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Moazeni-Roodi A, Aftabi S, Sarabandi S, et al. Genetic association between HOTAIR gene and the risk of cancer: an updated meta-analysis. J Genet. 2020;99:1–16. [PubMed] [Google Scholar]
- [22].Zeng L, Yang M, Gao D, et al. Prognostic and clinicopathological significance of long noncoding RNA GHET1 in human solid tumors: a meta-analysis. Clin Lab. 2020;66:7. [DOI] [PubMed] [Google Scholar]
- [23].Stroup DF, Berlin JA, Morton SC, et al. Meta-analysis of observational studies in epidemiology: a proposal for reporting. Meta-analysis Of Observational Studies in Epidemiology (MOOSE) group. JAMA. 2000;283:2008–12. [DOI] [PubMed] [Google Scholar]
- [24].Parmar MKB, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat Med. 1998;17:2815–34. [DOI] [PubMed] [Google Scholar]
- [25].Tierney JF, Stewart LA, Ghersi D, et al. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Nagy A, Lánczky A, Menyhárt O, et al. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep. 2018;8:9227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Higgins JP, Thompson SG, Deeks JJ, et al. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–48. [PubMed] [Google Scholar]
- [29].Dersimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–88. [DOI] [PubMed] [Google Scholar]
- [30].Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–101. [PubMed] [Google Scholar]
- [31].Egger M, Smith GD, Schneider M, et al. Bias in meta-analysis detected by a simple, graphical test. BMJ British Med J. 1998;316:469–71. [Google Scholar]
- [32].Fan Y, Wang YF, Su HF, et al. RETRACTED ARTICLE: Decreased expression of the long noncoding RNA LINC00261 indicate poor prognosis in gastric cancer and suppress gastric cancer metastasis by affecting the epithelial–mesenchymal transition. J Hematol Oncol. 2016;9:57. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [33].Xu TP, Huang MD, Xia R, et al. Decreased expression of the long non-coding RNA FENDRR is associated with poor prognosis in gastric cancer and FENDRR regulates gastric cancer cell metastasis by affecting fibronectin1 expression. J Hematol Oncol. 2014;7:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Zeng S, Xie X, Xiao YF, et al. Long noncoding RNA LINC00675 enhances phosphorylation of vimentin on Ser83 to suppress gastric cancer progression. Cancer Lett. 2018;412:179–87. [DOI] [PubMed] [Google Scholar]
- [35].Sun TT, He J, Liang Q, et al. A novel lncRNA GClnc1 promotes gastric carcinogenesis and may act as a modular scaffold of WDR5 and KAT2A complexes to specify the histone modification pattern. Cancer Discov. 2016;6:784–801. [DOI] [PubMed] [Google Scholar]
- [36].Sun T. The role of TMEFF2 and long non-coding RNA GAS1 in gastric carcinoma. Shanghai Jiao Tong University, 2015. [Google Scholar]
- [37].Liu Y, Sun M, Xia R, et al. LincHOTAIR epigenetically silences miR34a by binding to PRC2 to promote the epithelial-to-mesenchymal transition in human gastric cancer. Cell Death Dis. 2015;6:e1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Endo H, Shiroki T, Nakagawa T, et al. Enhanced expression of long non-coding RNA HOTAIR is associated with the development of gastric cancer. PLoS One. 2013;8:e77070e77070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Sun J. The clinical significance and molecular mechanism of long non-conding RNAs AC138128.1RP11-119F7.4 in gastric cancer. China Medical University (Liaoning), 2016. [Google Scholar]
- [40].Okugawa Y, Toiyama Y, Hur K, et al. Metastasis-associated long non-coding RNA drives gastric cancer development and promotes peritoneal metastasis. Carcinogenesis. 2014;35:2731–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Wu SW, Hao YP, Qui JH, et al. High expression of long noncoding RNA CCAT2 indicates poor prognosis of gastric cancer and promotes cell proliferation and invasion. Minerva Med. 2017;108:317–23. [DOI] [PubMed] [Google Scholar]
- [42].Peng W, Wu J, Fan H, et al. LncRNA EGOT promotes tumorigenesis via Hedgehog pathway in gastric cancer. Pathol Oncol Res Por. 2019;25:883–7. [DOI] [PubMed] [Google Scholar]
- [43].Zhao R, Zhang Y, Zhang X, et al. Exosomal long noncoding RNA HOTTIP as potential novel diagnostic and prognostic biomarker test for gastric cancer. Mol Cancer. 2018;17:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Zhang D, Sun G, Zhang H, et al. Long non-coding RNA ANRIL indicates a poor prognosis of cervical cancer and promotes carcinogenesis via PI3K/Akt pathways. Biomed Pharmacother. 2016;85:511. [DOI] [PubMed] [Google Scholar]
- [45].Gupta SC, Tripathi YN. Potential of long non‐coding RNAs in cancer patients: from biomarkers to therapeutic targets. Int J Cancer. 2017;140:1955–67. [DOI] [PubMed] [Google Scholar]
- [46].Kunej T, Obsteter J, Pogacar Z, et al. The decalog of long non-coding RNA involvement in cancer diagnosis and monitoring. Crit Rev Clin Lab Sci. 2014;51:344–57. [DOI] [PubMed] [Google Scholar]
- [47].Hung T, Chang HY. Long noncoding RNA in genome regulation: prospects and mechanisms. RNA Biol. 2010;7:582–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Li CY, Liang GY, Yao WZ, et al. Integrated analysis of long non-coding RNA competing interactions reveals the potential role in progression of human gastric cancer. Int J Oncol. 2016;48:1965–76. [DOI] [PubMed] [Google Scholar]
- [49].Liu Y, Jing Z, Zhang W, et al. lncRNA GAS5 enhances G1 cell cycle arrest via binding to YBX1 to regulate p21 expression in stomach cancer. Sci Rep. 2016;5:10159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Li L, Zhang L, Zhang Y, et al. Increased expression of LncRNA BANCR is associated with clinical progression and poor prognosis in gastric cancer. Biomed Pharmacother. 2015;72:109–12. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.