Abstract
The single-copy pahA gene from Penicillium chrysogenum encodes a phenylacetate 2-hydroxylase that catalyzes the first step of phenylacetate catabolism, an oxidative route that decreases the precursor availability for penicillin G biosynthesis. PahA protein is homologous to cytochrome P450 monooxygenases involved in the detoxification of xenobiotic compounds, with 84% identity to the Aspergillus nidulans homologue PhacA. Expression level of pahA displays an inverse correlation with the penicillin productivity of the strain and is subject to induction by phenylacetic acid. Gene expression studies have revealed a reduced oxidative activity of the protein encoded by pahA genes from penicillin-overproducing strains of P. chrysogenum compared to the activity conferred by phacA of A. nidulans. Sequencing and expression of wild-type pahA from P. chrysogenum NRRL 1951 revealed that an L181F mutation was responsible for the reduced function in present industrial strains. The mutation has been tracked down to Wisconsin 49–133, a mutant obtained at the Department of Botany of the University of Wisconsin in 1949, at the beginning of the development of the Wisconsin family of strains.
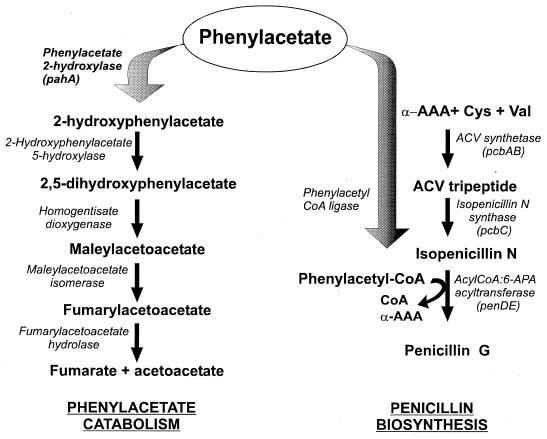
Phenylacetic acid (PA) is industrially used as a side chain precursor for the production of benzylpenicillin (penicillin G) by submerged fermentation of Penicillium chrysogenum. To incorporate PA into penicillin, the acid is activated as a coenzyme A thioester prior to the transacylation of the isopenicillin N precursor, substituting the l-α-aminoadipyl moiety by the phenylacetyl group. This acyl exchange is catalyzed by the acyl-coenzyme A (CoA):6-aminopenicillanic acid acyltransferase and constitutes the last step of penicillin G biosynthesis (Fig. 1). As this enzyme has broad substrate specificity, PA must be fed in excess to the cultures to promote the biosynthesis of penicillin G instead of other penicillins (12). The effectiveness of the side chain precursor appears to depend largely on its toxicity and its resistance to oxidation by P. chrysogenum (13). However, in industrial fermentation conditions, PA is not stoichiometrically channeled to penicillin G, and significant amounts of 2-hydroxyphenylacetic acid (2-OHPA) are detected in the broth. This suggests that a fraction of the precursor is oxidized by P. chrysogenum, and, from the point of view of economy, it would be interesting to ascertain this oxidative pathway in order to reduce PA losses. Rechanneling of the PA flux to the penicillin G biosynthetic pathway will probably reduce the amount of PA needed, lowering the cost of the fermentation process.
FIG. 1.
Competition between PA catabolism and penicillin G biosynthesis in P. chrysogenum. The PA catabolic pathway is the same proposed for A. nidulans (19). The enzyme phenylacetate 2-hydroxylase controls the incorporation of PA into a catabolic detoxification pathway, reducing the pool available for penicillin G biosynthesis. The genes known in P. chrysogenum are shown between brackets. From 2,5-dihydroxyphenylacetic or homogentisic acid, the PA catabolic pathway would be common to phenylalanine and tyrosine catabolism. α-AAA, α-aminoadipic acid; 6-APA, 6-aminopenicillanic acid; ACV, α-aminoadipyl-cysteinyl-valine.
The first reports on the specific catabolism of PA by microorganisms showed the presence of two alternative oxidative pathways, one through homoprotocatechuic acid and the other through homogentisic acid via 2-OHPA. The second one was shared by different microbes, including Pseudomonas fluorescens (18), Aspergillus niger (17), Nocardia salmonicolor (24), and also P. chrysogenum Wisconsin Q-176, in which crystals of 2-OHPA and homogentisic acid were isolated from culture filtrates (15, 20). Subsequent studies in other microorganisms detected the presence of 3-hydroxy- or 4-hydroxyphenylacetic acids instead of 2-OHPA as the first intermediates of this pathway. Nevertheless, homogentisic acid was isolated in all of them as a common intermediate before ring cleavage to give maleylacetoacetic acid, which was isomerized to fumarylacetoacetic acid. The last step of the pathway consisted in the hydrolysis of this compound, yielding fumaric and acetoacetic acids (Fig. 1), which can be incorporated into the Krebs cycle (26).
The ability of the ascomycete Aspergillus nidulans to grow on PA as the sole carbon source was the basis for the characterization of the catabolic pathway in this fungus (Fig. 1). The first step of this pathway involves a 2-hydroxylation of the PA ring by a microsomal cytochrome P450 [PA 2-hydroxylase; phenylacetate, NAD(P):oxygen oxidoreductase (2-hydroxylating); EC number 1.14.13], encoded by the phacA gene (phenylacetate catabolism) (19). The cytochrome P450 superfamily groups more than 500 hemoproteins that are widely distributed in nature. They catalyze diverse enzymatic reactions involved in the metabolism of endogenous compounds as sterols and vitamins together with the catabolism of toxic and xenobiotic compounds (21). Targeted disruption of the phacA gene in A. nidulans resulted in a partial inability to catabolyze PA by the transformants, together with a fivefold-increased penicillin G productivity (19). These results prompted us to characterize the homologous pahA (PA 2-hydroxylase) gene from P. chrysogenum and to study its influence on PA oxidation and penicillin G production.
MATERIALS AND METHODS
Fungal strains, media, and growth conditions.
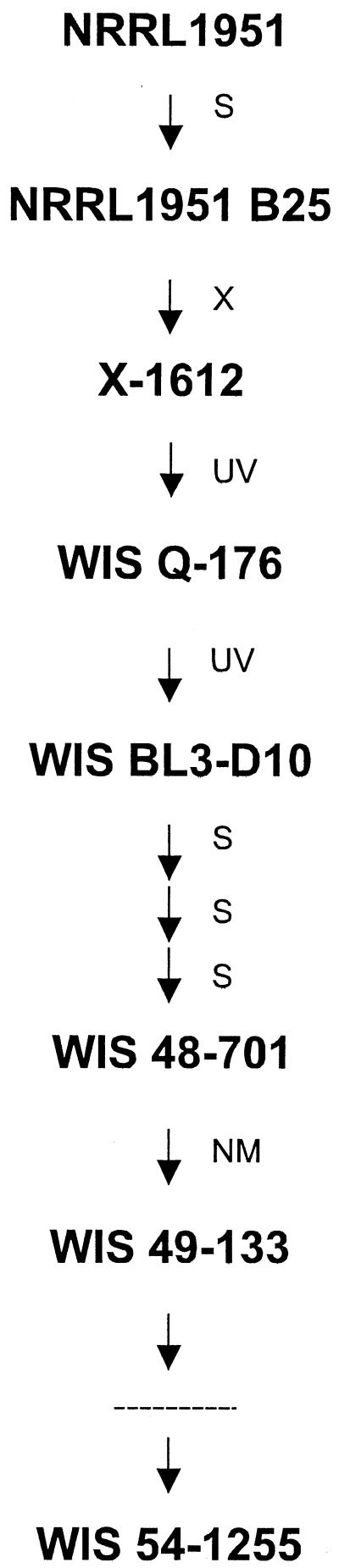
P. chrysogenum Wisconsin 54–1255 is a low-production strain widely employed in academic studies (3) and was the source of genomic DNA. The industrial strain P. chrysogenum E1, belonging to the Antibioticos S.A. series, was used for cDNA library construction. P. chrysogenum NRRL 1951 (ATCC 9480) is the ancestral wild-type strain isolated from a moldy cantaloupe at the Northern Regional Research Laboratories in Peoria, Ill., in 1943 (22). P. chrysogenum Wisconsin Q-176 (4) and Wisconsin 49–133 (3) are ancestors of Wisconsin 54–1255 (see Fig. 5). A. nidulans ATCC 32353 was used to test growth and resistance to PA. Growth of the fungi and penicillin production tests were performed as described (5).
FIG. 5.
Abbreviated genealogy of P. chrysogenum strains. S indicates selection without mutagenic treatment; X indicates selection following X irradiation; UV indicates selection following 275-nm UV irradiation; NM indicates selection following nitrogen mustard treatment. A dashed line between strains Wisconsin 49–133 and 54–1255 represents a total of five nitrogen mustard mutagenic steps followed by four selective steps linking both strains.
Bacterial strains, cloning vectors, and molecular biology techniques.
Escherichia coli DH5α (23) and E. coli TOP10 (Invitrogen) were the recipient strains for high-frequency transformation with vectors pBluescript I KS(+), pBC KS(+) (Stratagene), and pZERO-2 (Invitrogen). E. coli LE392 (23) and E. coli Sure (Stratagene) were the hosts of phage vectors λEMBL4 (23) and λZAP (Stratagene), respectively. Northern, Southern, and sequencing analyses were done by standard procedures (23).
PA assimilation and resistance.
PA assimilation was tested by seeding 1-μl aliquots containing 104 spores from each fungus on plates of Czapek-Dox minimal medium at pH 5.0, 6.0, or 7.0 with 0, 5, 10, 25, 50, or 100 mM PA as the sole carbon source. The resistance level was checked in the same media with 1% glucose and 0, 5, 10, 25, 50, or 100 mM PA. The plates were incubated at 25°C for 5 days.
PA and 2-OHPA oxidation tests.
Fungal cultures in 50 ml of PM medium (2) supplemented with 10% yeast extract were seeded with 107 spores and shaken at 250 rpm for 24 h at 25°C. Mycelium was recovered by filtration and washed with 0.9% NaCl. Wet mycelium (500 mg) was inoculated in 40 ml of PM without carbon source and shaken for 90 min before the addition of 2 g of PA or 2-OHPA per liter. Oxidation of both acids was followed by high-pressure liquid chromatography (HPLC) at 24-h intervals. Samples were diluted 10 times in 0.1 M sodium acetate (pH 7) (mobile phase) before injection in a Nucleosyl C18 column (250 by 4.6 mm; Waters) with a flow of 1 ml/min, detecting the elution of the samples by absorbance at 254 nm. Retention times were 16.5 min for PA and 21 min for 2-OHPA. Peaks corresponding to both acids were identified by coelution of standards (Sigma).
Cloning of the pahA gene.
A genomic library of P. chrysogenum Wisconsin 54–1255 constructed in λEMBL4 was screened using as a probe a 1.2-kb NotI cDNA fragment from the phacA gene of A. nidulans (19) according to standard procedures (23). Three recombinant phages were isolated and mapped, locating the hybridization signal in a 2.5-kb XhoI fragment that was subcloned in both orientations in pBluescript KS(+), giving pALP519 and pALP520. In order to clone the pahA gene from P. chrysogenum NRRL 1951, Wisconsin Q-176, and Wisconsin 49–133, total DNA from each strain was digested with XhoI, and fragments of 2 to 3 kb were purified by sucrose gradients and ligated into pZERO-2 (Invitrogen). After screening with a 1.2-kb EcoRV pahA probe from P. chrysogenum Wisconsin 54–1255, the pahA genes corresponding to the different strains were cloned as 2.5-kb XhoI fragments. The cDNA expression library was constructed with mRNA purified from stirred-tank fermentations of P. chrysogenum E1 in penicillin G production conditions (11). The expression library was screened with the 1.2-kb EcoRV fragment internal to the pahA gene from P. chrysogenum Wisconsin 54–1255 as the probe. After partial sequencing of the 5′ ends of 12 clones, the largest one (pALP555) was fully sequenced in both strands.
Fungal transformation systems.
P. chrysogenum was transformed by described protocols (9) using 4 mg/ml Caylase (Cayla, Toulouse, France) for protoplast formation. Phleomycin-resistant transformants were selected on Czapek-Dox minimal medium osmotically stabilized with 1 M sorbitol and supplemented with 30 μg/ml phleomycin. The fungal integrative vector pALfleo7 (10) was used for the construction of expression vectors. pALP956, containing the pahA gene from P. chrysogenum Wisconsin 54–1255, was constructed by subcloning a 4.1-kb BamHI-XhoI fragment in the EcoRV site of pALfleo7. pALP971 is a similar construction carrying the gene from P. chrysogenum NRRL 1951. pALP906 contains a 4.3-kb BamHI fragment including the phacA gene from A. nidulans.
Transcriptional analysis of pahA gene.
Mycelium samples were frozen in liquid N2, and total RNA was purified using RNeasy (Qiagen). Northern blots were hybridized using as probe the 1.2-kb EcoRV fragment internal to the pahA gene.
Nucleotide sequence accession numbers.
The nucleotide sequences of phenylacetate 2-hydroxylase genes from P. chrysogenum strains have been deposited in GenBank under accession numbers AF056978 (strain Wisconsin 54–1255) and AF057558 (strain NRRL 1951). The amino acid sequences of these proteins can be accessed through NCBI Protein Database under accession numbers AAF00011 (strain Wisconsin 54–1255) and AAF21759 (strain NRRL 1951).
RESULTS
P. chrysogenum is unable to use PA as sole carbon source.
To test the ability of P. chrysogenum Wisconsin 54–1255 and A. nidulans to utilize PA as a sole carbon source, aliquots of 104 conidia were seeded on plates of Czapek minimal medium where sucrose had been replaced with 0, 5, 10, 25, 50, or 100 mM PA. The experiment was carried out at pH 5.0, 6.0, or 7.0 due to the pH-dependent PA toxicity (12). Residual growth in the absence of a carbon source was attributed to slow assimilation of the agar and was independent of the pH of the medium. PA toxicity prevented residual growth at pH 5.0, whereas at pH 6.0 the growth inhibition was reduced, and at pH 7.0, inhibition disappeared. While A. nidulans showed enhanced growth over the control without a carbon source at several PA concentrations, this was not the case for P. chrysogenum Wisconsin 54–1255, which showed the same residual growth as the control without a carbon source or no growth at all. Finally, the wild-type strain P. chrysogenum NRRL 1951 and the industrial strain E1 were tested in the same conditions. The behavior of NRRL 1951 was very similar to that of strain Wisconsin 54–1255, whereas the industrial strain E1 showed no residual growth at any PA concentration. These nutritional differences directed us to the characterization of the P. chrysogenum gene homologous to A. nidulans phacA.
Resistance of P. chrysogenum to PA.
In order to optimize a PA resistance test, the MIC of this acid was tested for P. chrysogenum and A. nidulans in Czapek minimal medium with 1% glucose as the main carbon source and PA at concentrations ranging from 0 to 100 mM. As the MIC was expected to be pH dependent, three different PA ranges were prepared, pH 5.0, 6.0, and 7.0. The results confirmed higher toxicity at pH 5.0. Having an alternative carbon source, A. nidulans and P. chrysogenum NRRL 1951 showed similar MICs: 25 mM at pH 5.0, >100 mM at pH 6.0, and no change at pH 7.0. Improved strains Wisconsin 54–1255 and E1 were progressively less resistant; the MICs were 10 mM and <5 mM, respectively, at pH 5.0 and 100 mM and 25 mM, respectively, at pH 6.0. At pH 7.0 growth of Wisconsin 54–1255 was unaffected by PA concentration, but strain E1 showed a progressive growth reduction with increasing PA, being almost totally inhibited at 100 mM. These results indicate an inverse correlation between PA resistance and penicillin productivity.
Characterization of pahA gene from P. chrysogenum Wisconsin 54–1255 and industrial strain E1.
The pahA gene from Wisconsin 54–1255 was isolated by hybridization of a genomic library with a 1.2-kb NotI probe corresponding to the A. nidulans homologue phacA, which encodes an enzyme with PA 2-hydroxylase activity (19). Sequence analysis of a 2.5-kb XhoI fragment hybridizing to the phacA probe with the codon preference algorithm (Geneplot, DNASTAR) showed a coding region of 1,723 nucleotides interrupted by three introns of 57, 64, and 55 bp. The existence of the predicted introns was verified by comparison with the open reading frame (ORF) of 1,548 nucleotides found in the cDNA sequence of plasmid pALP555 isolated from a P. chrysogenum E1 expression library. A single-nucleotide substitution located 35 bp downstream from the translation stop codon was found between genomic and cDNA sequences. This nucleotide change, verified by sequencing additional genomic and cDNA clones, does not affect the protein sequence but could affect transcriptional efficiency or mRNA stability. Probably, this base substitution reflects the 45 years of divergence in the strain improvement programs between strains Wisconsin 54–1255 and E1, used to construct the genomic and expression libraries, respectively.
The deduced protein corresponding to the enzyme PA 2-hydroxylase (PahA) was 516 residues long and had a predicted molecular mass of 58.1 kDa. The protein sequence was 84% identical to that of the A. nidulans homologue PhacA (NCBI Protein Database accession no. CAB43093) (19), and database searches revealed that they are homologous to cytochrome P450 monooxygenases, involved in the detoxification of xenobiotic compounds. The cysteine heme-iron ligand signature FW-SGNH-x-GD-x-RKHPT-x-C-LIVMFAP-GAD (Prosite database entry PS00086), characteristic of the cytochrome P450 protein family, was conserved in PahA and PhacA except for the Phe residue, which was conservatively replaced with Tyr. According to the P450 nomenclature, PahA would be designated CYP504 in the new family defined for the A. nidulans homologue PhacA (19).
Regulation of transcription of pahA gene.
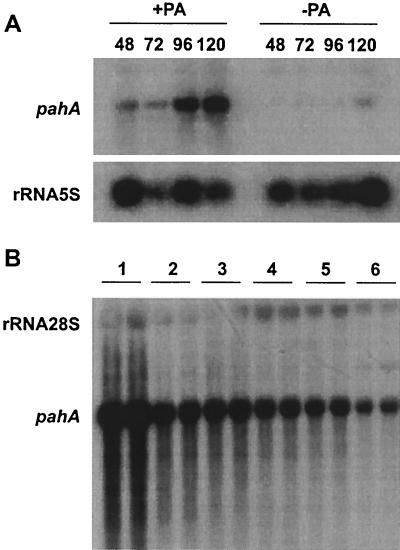
The transcription of the pahA gene was followed and quantified along penicillin fermentations by Northern hybridization. Flask fermentations were carried out in penicillin production medium (5) in the presence and absence of PA (4 g/liter). Maximal expression level was reached in the presence of PA, corresponding to the standard fermentation medium. The absence of PA resulted in a strong decrease (around 75%) in the transcription level of pahA (Fig. 2A). The expression level of pahA was also analyzed in a series of P. chrysogenum strains with different penicillin productivities. An inverse correlation was found between penicillin productivity and pahA gene expression in the series (Fig. 2B). This inverse correlation was consistent with the results of the PA resistance tests in solid medium.
FIG. 2.
Expression of pahA gene. (A) Expression of pahA was detected by Northern blot using total RNA isolated from P. chrysogenum E1 at 48, 72, 96, and 120 h of fermentation in standard penicillin G production conditions with and without PA. In the standard medium (with PA), expression is correlated with the penicillin production kinetics. The 5S rRNA signal was used as a loading control. (B) Northern hybridization of pahA with different P. chrysogenum strains grown in penicillin G production conditions. Duplicate lanes were loaded with total RNA from each of the following strains: lane 1, Wisconsin 54–1255; lanes 2 to 5, industrial strains with increasing penicillin productivity from left to right; lane 6, strain E1. The size of the pahA transcript corresponds to 1.6 kb.
PahA from improved P. chrysogenum strains displays reduced oxidative activity when compared to the A. nidulans homologue.
The differences in nutritional behavior shown by P. chrysogenum and A. nidulans in the utilization of PA as a sole carbon source suggested a possible impairment of its catabolic pathway in P. chrysogenum. To test this hypothesis, we compared the PahA activities of the enzymes encoded by pahA from P. chrysogenum Wisconsin 54–1255 and phacA from A. nidulans expressed in transformants of P. chrysogenum Wisconsin 54–1255. The expression vectors pALP906 (phacA from A. nidulans) and pALP956 (pahA from Wisconsin 54–1255) were constructed, and single-copy transformants of P. chrysogenum Wisconsin 54–1255 were selected by Southern blotting. PA assimilation in solid medium revealed the ability to use this compound as a sole carbon source only by those transformants carrying the phacA gene.
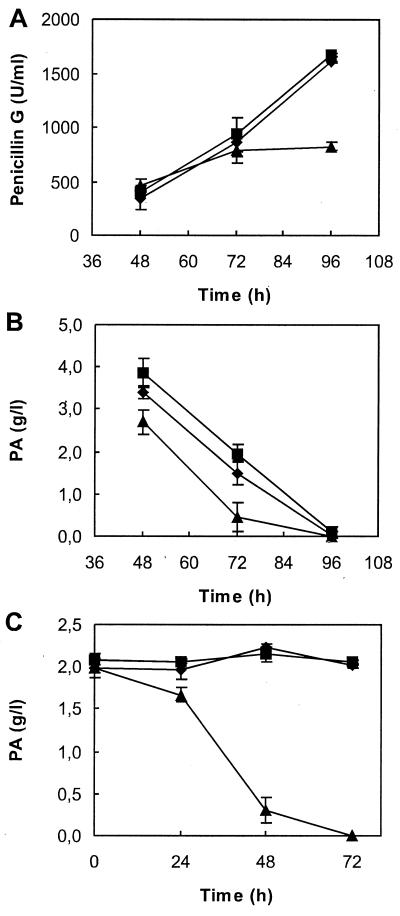
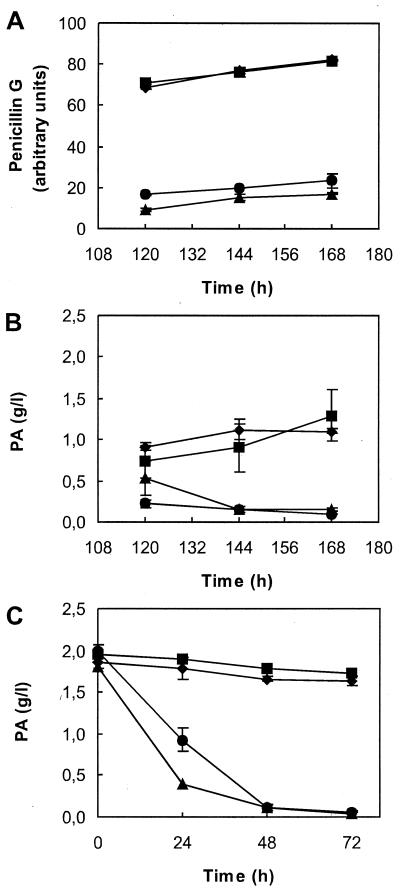
In order to confirm these results, transformants T-06 (pALP956) and T-16 (pALP906) were selected to test penicillin G production and PA oxidation in flask fermentations (Fig. 3). Whereas no significant differences were found in penicillin productivity or PA oxidation between T-06 (pahA) and the parental untransformed strain P. chrysogenum Wisconsin 54–1255, T-16 (phacA) showed 50% lower penicillin G productivity (Fig. 3A) and a considerable increase in the PA consumption rate during the fermentation: the PA was almost exhausted at 72 h, 24 h earlier than in the parental strain or T-06 (Fig. 3B). The PA oxidation test showed no significant PA oxidative activity in P. chrysogenum Wisconsin 54–1255 or T-06, whereas T-16 showed a clear PA oxidative activity, which was totally consumed at 72 h of incubation (Fig. 3C). The oxidative activity on 2-OHPA followed a similar pattern to PA (not shown), indicating that phacA is involved in the catabolism of both acids.
FIG. 3.
Phenotype of transformants of P. chrysogenum Wisconsin 54–1255 expressing pahA and phacA gene constructions. The analysis was carried out with the untransformed strain Wisconsin 54–1255 (diamonds), transformant T-06 carrying an extra copy of endogenous pahA (squares), and transformant T-16 carrying phacA from A. nidulans (triangles). (A and B) Penicillin G production and PA consumption in penicillin fermentation conditions. Flask fermentations were carried out in standard conditions measuring penicillin G production and PA consumption by HPLC at the time points indicated. Transformant T-16, carrying phacA, showed a strong reduction in penicillin production and enhanced PA consumption. (C) PA oxidation test in liquid minimal medium. Mycelium of each strain was grown on glucose-supplemented medium for 24 h before transfer to minimal medium with PA as the sole carbon source. Flasks were shaken at 25°C, and samples were taken at the time points indicated to quantify the remaining PA by HPLC. Again, only T-16 showed capacity to consume PA. Vertical bars indicate standard deviations.
PahA activity in wild-type strain P. chrysogenum NRRL 1951.
A possible explanation for the reduced activity of the PahA encoded by the pahA gene of P. chrysogenum improved strains could be that, during the industrial strain development programs, a mutant with reduced PahA function had been selected in the ancestral phylogeny shared by strains Wisconsin 54–1255 and E1. In principle, the nutritional tests pointed against this possibility: the wild-type strain P. chrysogenum NRRL 1951 was also unable to use PA as a carbon source in the conditions tested, behaving in this aspect like the industrial strains. The loss-of-function mutation hypothesis could be easily ascertained by comparison of the sequence of pahA isolated from improved strains to the sequence of the wild-type gene. This gene, isolated as a 2.5-kb XhoI fragment from strain NRRL 1951, was sequenced and subcloned for the construction of expression vector pALP971. Sequence comparisons of the pahA genes showed a base change at position 598 of the ORF, where the T present in strains Wisconsin 54–1255 and E1 was formerly a C in wild-type strain NRRL 1951. The mutation C598 → T causes a single-amino-acid substitution at position 181 of the protein: a leucine residue in the wild-type strain has been substituted with phenylalanine in the improved strains (L181F). This amino acid change is otherwise located in a highly conserved region compared to the sequences of A. nidulans PhacA and a cytochrome P450 monooxygenase from Aspergillus terreus (NCBI Protein Database accession no. AAD34565) (16). Both of them agree with the leucine of wild-type P. chrysogenum NRRL 1951.
The biological significance of the mutation was checked by the expression in the industrial strain E1 of the pahA genes from NRRL 1951 and Wisconsin 54–1255 and the phacA gene from A. nidulans. The transformants were tested for PA assimilation, and as expected, those carrying pahA genes from NRRL 1951 or Wisconsin 54–1255 were unable to grow on PA as a sole carbon source, while those carrying phacA showed enhanced growth on PA. Single-copy transformants T-49 (phacA), T-50 (pahA from Wisconsin 54–1255), and T-93 (pahA from NRRL 1951) were selected by Southern hybridization and tested for penicillin G production and PA oxidation (Fig. 4). Surprisingly, the pahA gene from NRRL 1951 conferred on E1 transformants the ability to oxidize PA, even though at a lower level than the phacA gene. In penicillin G flask fermentations, production decreased about 80% in T-49 (phacA) and 70% in T-93 (wild-type pahA) compared to the untransformed industrial strain E1 (Fig. 4A). Both T-49 and T-93 transformants displayed a reduced PA level in the fermentation broth (Fig. 4B). In PA oxidation tests, the difference was clear after 24 h, when T-49 had assimilated about 80% of initial PA and T-93 about 50%, whereas T-50 and the untransformed strain E1 showed only a slight oxidation of this compound (Fig. 4C). Similar differences were also found in the assimilation of 2-OHPA (not shown). These results indicated that the base substitution at position 598 of the pahA gene from strains Wisconsin 54–1255 and E1 is a loss-of-function mutation selected in a common ancestor of both strains as a mutation enhancing penicillin production.
FIG. 4.
Phenotype of transformants of P. chrysogenum E1 expressing phacA from A. nidulans, pahA from P. chrysogenum Wisconsin 54–1255, and pahA from P. chrysogenum NRRL 1951. The analysis was carried out with the untransformed strain E1 (diamonds), transformant T-49 (phacA, triangles), transformant T-50 (pahA from Wisconsin 54–1255, squares), and transformant T-93 (pahA from NRRL 1951, circles). (A) Penicillin G production and (B) PA consumption in penicillin fermentation conditions. Flask fermentations were carried out with daily additions of 1 g of PA per liter, measuring penicillin G production and PA consumption by HPLC at the time points indicated. Transformants T-49 and T-93 showed reduced penicillin G production and enhanced PA consumption. (C) PA oxidation test in liquid minimal medium with PA as the sole carbon source. Both T-49 and T-93 showed enhanced capacity to consume this compound. Vertical bars indicate standard deviations.
Location of loss-of-function mutation of pahA in the phylogeny of P. chrysogenum improved strains.
In order to locate the mutational step that gave rise to pahA inactivation, two strains from the early ancestry of the Wisconsin family were chosen (Fig. 5). The first one, P. chrysogenum Wisconsin Q-176, represents the origin of the Wisconsin family and was obtained by 275-nm UV irradiation of spores from strain X-1612. Tested in 300-liter fermentors, strain Q-176 gave yields of over 900 U/ml, in contrast to about 500 U/ml obtained with strain X-1612 under the same conditions (4). The second strain chosen, Wisconsin 49–133, is located five steps down from Wisconsin Q-176. First, a UV mutagenic treatment of Q-176 was devoted to the selection of the pigmentless strain BL3-D10. This strain was selected to eliminate the yellow pigment naturally produced by P. chrysogenum in order to get a white commercial product, avoiding antibiotic losses during the extraction of the pigment, but involved a 25% reduction in penicillin yield (3). Three successive selection rounds without mutagenic treatment of BL3-D10 gave strains 47–638, 47–1564, and 48–701, respectively, the last one with a production level 15% above that of Q-176. Wisconsin 49–133 was obtained by nitrogen mustard mutagenic treatment of strain 48–701 and represented the greatest single improvement among the Wisconsin pigmentless series, with penicillin yields 100% above that of the 48–701 parent, only 50% as much mycelium, and an “exceptionally efficient utilization of PA” (1). This particular description is consistent with the phenotype expected for the pahA loss-of-function mutation. The sequence analysis of the pahA genes from strains Q-176 and 49–133 revealed that, while a C was found at position 598 of the gene from strain Q-176 (similar to the ancestor NRRL 1951), it had been replaced with a T in strain 49–133 (similar to its descendant Wisconsin 54–1255). The presence of the pahA mutation in strain Wisconsin 49–133 could account for its improvement in PA utilization for penicillin production, with a growth reduction probably due to the increased PA toxicity in the absence of the PA oxidative activity encoded by pahA.
DISCUSSION
The toxicity of PA is based on its properties as a weak acid. At low pH, the undissociated species can diffuse across the lipid bilayer into the cell (13), where a higher cytoplasmic pH causes the protonation of the molecules and their intracellular accumulation. This accumulation causes the dissipation of the pH gradient, affecting transport systems and producing serious damage to the cell (14).
In penicillin-producing fungi, the toxicity of PA can be avoided by two competing mechanisms, catabolic degradation and detoxification by incorporation into penicillin. The second mechanism has been proposed as an example of the detoxification hypothesis for the origin of several antibiotic biosynthesis pathways (8). According to this scheme, the inactivation of the PA catabolic pathway in A. nidulans resulted in a fivefold increase in penicillin titer (19). The results of the nutritional tests on PA assimilation performed in this work have shown a clear phenotypic difference between A. nidulans and P. chrysogenum: all the P. chrysogenum strains tested were unable to use PA as a sole carbon source, suggesting a block in the catabolic pathway. These results indicate that the two abovementioned alternative detoxification mechanisms could be oppositely balanced in both species, with catabolic detoxification predominating in A. nidulans and penicillin detoxification in P. chrysogenum. According to this detoxification hypothesis, resistance to PA has been used for the screening of P. chrysogenum mutants with improved penicillin productivity (7). Nevertheless, our PA resistance tests pointed to an inverse correlation between PA resistance and penicillin productivity.
The pahA gene encodes a cytochrome P450 that shows 84% identity with the A. nidulans homologue PhacA. Comparison of PahA and PhacA activities by gene expression studies showed that the block in PA catabolism found in P. chrysogenum could be originated by inactivation or strong reduction of PahA activity. Transformants of P. chrysogenum carrying the A. nidulans phacA gene showed (i) assimilation of PA as the sole carbon source on solid medium, (ii) increased resistance to PA in the presence of an alternative carbon source, (iii) ability to oxidize PA and 2-OHPA in liquid medium, and (iv) reduced penicillin G titer in flask fermentations in the presence of PA as a side chain precursor. The reduced enzymatic activity of PahA in penicillin-overproducing strains of P. chrysogenum seems to be due to an L181F mutation introduced during the strain improvement programs. The pahA gene from wild-type strain NRRL 1951, lacking this mutation, conferred a phenotype very similar to A. nidulans phacA.
The study of the genealogy of the Wisconsin family of P. chrysogenum pointed to Wisconsin 49–133 as a strain with phenotypic descriptions expected for a pahA loss-of-function mutation: double penicillin titer with 50% less growth and much better efficiency of PA utilization for penicillin production, especially at higher precursor levels (1). The analysis of the pahA sequence of this strain revealed the presence of the L181F mutation, while its ancestor Wisconsin Q-176 carried a wild-type pahA, similar to strain NRRL 1951. As strains Q-176 and 49–133 were only two mutagenic treatments apart, and the first one was devoted to the isolation of a pigmentless strain with a reduction in productivity, the loss-of-function mutation of pahA should have occurred at the second mutagenic step. This was the first nitrogen mustard mutagenesis carried out in the Wisconsin series, an experiment that gave rise to strain 49–133 in 1949. This conclusion is also supported by the “exceptional efficiency in the PA utilization for penicillin production” outlined for strain 49–133 (1), the phenotype expected for the pahA loss-of-function mutation.
Although a negative correlation of PahA activity and penicillin G productivity is clearly supported by the results presented here, some open questions remain. If PahA activity was totally blocked in P. chrysogenum industrial strains, then the origin of 2-OHPA in the fermentation broth would still be unexplained. Either the 2-OHPA could be produced by nonspecific oxidative enzymatic activities, or, alternatively, the blockage of PahA could be partial. Some results could be consistent with this second hypothesis: low but regulated transcription levels of pahA were found even in the highest producers, with an inverse correlation between transcription level and penicillin productivity. Maybe a low percentage of activity, undetectable in the oxidation tests, has been preserved to sustain growth in the PA-fed fermentors and is enough to accumulate the 2-OHPA. Another surprising result is the contradictory behavior of strain NRRL 1951, unable to assimilate PA on solid medium but with a level of PA oxidative activity in liquid cultures comparable to that of A. nidulans.
The dysfunction of PA assimilation could have been one of the most important genetic improvements of the strains currently used for the commercial production of penicillin, pairing with the selection of improved strains with increased copy number of the penicillin biosynthesis gene cluster (6, 25). After 60 years of classical mutation and screening programs, current P. chrysogenum industrial strains constitute one of the best resources to validate metabolic engineering design, though being, on the other hand, hard to improve with these directed approaches.
ACKNOWLEDGMENTS
We thank J. A. González, P. Merino, M. Sandoval, and M. T. García for their excellent technical assistance and A. T. Marcos and J. Velasco for helpful comments.
M. Rodríguez-Sáiz was supported by a fellowship from the Spanish Ministry of Education and Science.
REFERENCES
- 1.Anderson R F, Whitmore L M, Brown W E, Peterson W H, Churchill B W, Roegner F R, Campbell T H, Backus M P, Stauffer J F. Production of penicillin by some pigmentless mutants of the mold, Penicillium chrysogenumQ176. Ind Eng Chem. 1953;45:768–773. [Google Scholar]
- 2.Anné J. Somatic hybridization between Penicillium chrysogenumspecies after induced fusion of their protoplasts. Agricultura. 1977;25:1–117. [Google Scholar]
- 3.Backus M P, Stauffer J F. The production and selection of a family of strains in Penicillium chrysogenum. Mycologia. 1955;47:429–463. [Google Scholar]
- 4.Backus M P, Stauffer J F, Johnson M J. Penicillin yields from new mold strains. J Am Chem Soc. 1946;68:152–153. doi: 10.1021/ja01205a518. [DOI] [PubMed] [Google Scholar]
- 5.Barredo J L, Alvarez E, Cantoral J M, Díez B, Martín J F. Glucokinase-deficient mutant of Penicillium chrysogenumis derepressed in glucose catabolite regulation of both β-galactosidase and penicillin biosynthesis. Antimicrob Agents Chemother. 1988;32:1061–1067. doi: 10.1128/aac.32.7.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barredo J L, Díez B, Alvarez E, Martín J F. Large amplification of a 35 kb DNA fragment carrying two penicillin biosynthetic genes in high penicillin producing strains of Penicillium chrysogenum. Curr Genet. 1989;16:453–459. doi: 10.1007/BF00340725. [DOI] [PubMed] [Google Scholar]
- 7.Barrios-González J, Montenegro E, Martín J F. Penicillin production by mutants resistant to phenylacetic acid. J Ferment Bioeng. 1993;76:455–458. [Google Scholar]
- 8.Dhar M M, Khan A W. Formation of antibiotics. Nature. 1971;233:182–184. doi: 10.1038/233182a0. [DOI] [PubMed] [Google Scholar]
- 9.Díez B, Alvarez E, Cantoral J M, Barredo J L, Martín J F. Selection and characterization of pyrG mutants of Penicillium chrysogenum lacking orotidine-5′-phosphate decarboxylase and complementation by the pyr4 gene of Neurospora crassa. Curr Genet. 1987;12:277–282. [Google Scholar]
- 10.Díez B, Mellado E, Rodríguez M, Bernasconi E, Barredo J L. The NADP-dependent glutamate dehydrogenase gene from Penicillium chrysogenumand the construction of expression vectors for filamentous fungi. Appl Microbiol Biotechnol. 1999;52:196–207. doi: 10.1007/s002530051509. [DOI] [PubMed] [Google Scholar]
- 11.Díez B, Schleissner C, Moreno M A, Rodríguez M, Collados A, Barredo J L. The manganese superoxide dismutase from the penicillin producer Penicillium chrysogenum. Curr Genet. 1998;33:387–394. doi: 10.1007/s002940050351. [DOI] [PubMed] [Google Scholar]
- 12.Hersbach G J M, Van Der Beek C P, Van Dijck P W M. The penicillins: properties, biosynthesis and fermentation. In: Vandamme E J, editor. Biotechnology of industrial antibiotics. Vol. 3. New York, N.Y: Marcel Dekker Inc.; 1984. pp. 45–140. [Google Scholar]
- 13.Hillenga D J, Versantvoort H J, van der Molen S, Driessen A J, Konings W N. Penicillium chrysogenumtakes up the penicillin G precursor phenylacetic acid by simple passive diffusion. Appl Environ Microbiol. 1995;61:2589–2595. doi: 10.1128/aem.61.7.2589-2595.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hunter D R, Segel H. Effect of weak acids on amino acid transport by Penicillium chrysogenum: evidence for a proton or charge gradient as the driving force. J Bacteriol. 1973;113:1184–1192. doi: 10.1128/jb.113.3.1184-1192.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Isono M. Studies on the oxidation of phenylacetic acid by Penicillium chrysogenumQ 176. J Agric Chem Jpn. 1953;27:297–301. [Google Scholar]
- 16.Kennedy J, Auclair K, Kendrew S G, Park C, Vederas J C, Hutchinson C R. Modulation of polyketide synthase activity by accessory proteins during lovastatin biosynthesis. Science. 1999;284:1368–1372. doi: 10.1126/science.284.5418.1368. [DOI] [PubMed] [Google Scholar]
- 17.Kluyver A J, van Zijp J C M. The production of homogentisic acid out of phenylacetic acid by Aspergillus niger. Antonie van Leeuwenhoek. 1951;17:47–55. doi: 10.1007/BF02062278. [DOI] [PubMed] [Google Scholar]
- 18.Kunita N. Bacterial oxidation of phenylacetic acid. II. The pathway through homogentisic acid. Med J Osaka Univ. 1955;3:703–708. [Google Scholar]
- 19.Mingot J M, Peñalva M A, Fernández-Cañón J M. Disruption of phacA, an Aspergillus nidulansgene encoding a novel cytochrome P450 monooxygenase catalyzing phenylacetate 2-hydroxylation, results in penicillin overproduction. J Biol Chem. 1999;274:14545–14550. doi: 10.1074/jbc.274.21.14545. [DOI] [PubMed] [Google Scholar]
- 20.Nishikida T. Isolation of o-hydroxyphenylacetic acid from the penicillin impurities. J Antibiot. 1951;4:299–300. [Google Scholar]
- 21.Porter T D, Coon M J. Cytochrome P-450: multiplicity of isoforms, substrates, and catalytic and regulatory mechanisms. J Biol Chem. 1991;266:13469–13472. [PubMed] [Google Scholar]
- 22.Raper K B, Alexander D F. Penicillin. V. Mycological aspects of penicillin production. J Elisha Mitchell Sci Soc. 1945;61:74–113. [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 24.Sariaslani F S, Harper D B, Higgins I J. Microbial degradation of hydrocarbons: catabolism of 1-phenylalkanes by Nocardia salmonicolor. Biochem J. 1974;140:31–45. doi: 10.1042/bj1400031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith D J, Bull J H, Edwards J, Turner G. Amplification of the isopenicillin N synthetase gene in a strain of Penicillium chrysogenumproducing high levels of penicillin. Mol Gen Genet. 1989;216:492–497. doi: 10.1007/BF00334395. [DOI] [PubMed] [Google Scholar]
- 26.Van den Tweel W J J, Smits J P, Nbont J A M. Catabolism of dl-α-phenylhydrocrylic, phenylacetic and 4-hydroxyphenylacetic via homogentisic acid into a Flavobacteriumsp. Arch Microbiol. 1988;149:207–213. [Google Scholar]