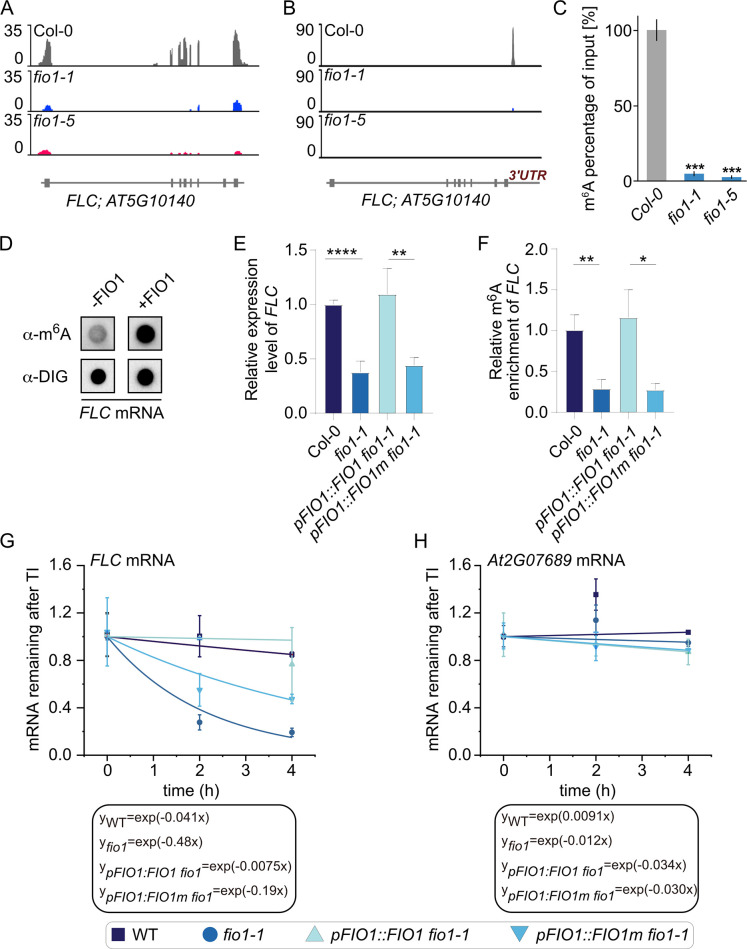
Fig 6. FIONA1 acts as m6A-methyltransferase on FLC.
(A) RNA-seq coverage observed at the FLC locus. RNA-seq reads in Col-0 (grey), fio1-1 (blue) and fio1-3 (pink). Gene model depicts exons and introns. (B) MeRIP-seq coverage observed at the FLC locus. RNA-seq reads in Col-0 (grey), fio1-1 (blue) and fio1-3 (pink). Gene model depicts exons and introns. (C) Percentages of the m6A methylated FLC mRNA in input samples in the wild type, fio1-1 and fio1-3 measured by m6A-IP-qRT PCR. Values are the means ±SD. N = 4, ***P≤ 0.001. (D) In vitro methylation of FLC mRNA. FIO1 was purified as GST-tagged protein from E. coli and incubated with digoxygenin (DIG)-labeled in vitro produced FLC mRNA in the presence of SAM. Dot blot in the upper row show m6A methylated signal with the anti- m6A antibody. The dots in the lower row the signal of the anti-DIG antibody. Increased signal intensities were detected when FIO1 enzyme was present. (E) qPCR results showing the relative expression of FLC in 12-day-old Col-0, fio1, pFIO1:FIO1/fio1, and pFIO1:FIO1m/fio1 seedlings. Data are means ± SD for 3 biological replicates × 3 technical replicates. * p < 0.05, ** p < 0.01 by t test (two-tailed). (F) m6A-IP-qPCR results showing the relative m6A levels of FLC transcripts in 12-day-old Col-0, fio1, pFIO1:FIO1/fio1, and pFIO1:FIO1m/fio1 seedlings. Data are means ± SD for 3 biological replicates × 3 technical replicates. * p < 0.05, ** p < 0.01 by t test (two-tailed). (G) and (H) The mRNA lifetimes of FLC (G) in Col-0, fio1, pFIO1:FIO1/fio1, and pFIO1:FIO1m/fio1. The AT2G07689 (H) was used as the negative control. TI: transcription inhibition. Data are represented as means ± SD for 2 biological replicates × 3 technical replicates.