Summary
The environmental bacterium Legionella pneumophila causes the pneumonia Legionnaires' disease. The opportunistic pathogen forms biofilms and employs the Icm/Dot type IV secretion system (T4SS) to replicate in amoebae and macrophages. A regulatory network comprising the Legionella quorum sensing (Lqs) system and the transcription factor LvbR controls bacterial motility, virulence and biofilm architecture. Here we show by comparative proteomics that in biofilms formed by the L. pneumophila ΔlqsR or ΔlvbR regulatory mutants the abundance of proteins encoded by a genomic ‘fitness island’, metabolic enzymes, effector proteins and flagellar components (e.g. FlaA) varies. ∆lqsR or ∆flaA mutants form ‘patchy’ biofilms like the parental strain JR32, while ∆lvbR forms a ‘mat‐like’ biofilm. Acanthamoeba castellanii amoebae migrated more slowly through biofilms of L. pneumophila lacking lqsR, lvbR, flaA, a functional Icm/Dot T4SS (∆icmT), or secreted effector proteins. Clusters of bacteria decorated amoebae in JR32, ∆lvbR or ∆icmT biofilms but not in ∆lqsR or ∆flaA biofilms. The amoeba‐adherent bacteria induced promoters implicated in motility (P flaA ) or virulence (P sidC , P ralF ). Taken together, the Lqs‐LvbR network (quorum sensing), FlaA (motility) and the Icm/Dot T4SS (virulence) regulate migration of A. castellanii through L. pneumophila biofilms, and – apart from the T4SS – govern bacterial cluster formation on the amoebae.
Abbreviations
- Icm/Dot
intracellular multiplication/defective organelle trafficking;
- LAI‐1
Legionella autoinducer‐1;
- LCV
Legionella‐containing vacuole;
- Lqs
Legionella quorum sensing;
- LvbR
Legionella virulence and biofilm regulator;
- c‐di‐GMP
cyclic di‐guanosine monophosphate;
- GFP
green fluorescent protein;
- T4SS
type IV secretion system
Introduction
Legionella pneumophila is a Gram‐negative environmental bacterium that resides in freshwater niches. Through inhalation of L. pneumophila‐laden aerosols the opportunistic pathogen reaches the lung and infects alveolar macrophages, which can cause the life‐threatening Legionnaires' disease (Fields et al., 2002; Newton et al., 2010; Hilbi et al., 2011). Legionella pneumophila replicates intracellularly within free‐living protozoa, which appear to represent the preferential niche in the environment (Fields, 1996; Murga et al., 2001; Kuiper et al., 2004). The pathogen governs the interactions with host cells through the Icm/Dot type IV secretion system (T4SS) (Kubori and Nagai, 2016; Ghosal et al., 2019; Böck et al., 2021), which transports the impressive number of >300 different ‘effector’ proteins implicated in the formation of a unique, ER‐associated compartment, the Legionella‐containing vacuole (LCV) (Isberg et al., 2009; Hubber and Roy, 2010; Finsel and Hilbi, 2015; Personnic et al., 2016; Qiu and Luo, 2017; Steiner et al., 2018).
Legionella pneumophila is a facultative intracellular bacterium, which also colonizes and persists in complex biofilms composed of various microorganisms, including bacteria and protozoa (Lau and Ashbolt, 2009; Declerck, 2010; Abdel‐Nour et al., 2013; Hoffmann et al., 2014; Swart et al., 2018). Predatory protozoa, such as the amoeba Acanthamoeba castellanii, migrate and ‘graze’ on the bacterial biofilm communities (Huws et al., 2005). Intriguingly, amoeba migration and endocytic processes such as micropinocytosis are antagonistically correlated, presumably, since they largely require the same cellular machinery implicated in cytoskeleton and membrane dynamics (Veltman, 2015; Delgado et al., 2022). Intracellular replication within amoebae seems to promote bacterial features that facilitate biofilm formation by L. pneumophila (Bigot et al., 2013) and is likely important for L. pneumophila to grow within multispecies biofilms (Declerck et al., 2007; Declerck et al., 2009).
Under laboratory conditions, L. pneumophila forms mono‐species biofilms that allow the investigation of bacterial determinants potentially implicated in biofilm development (Mampel et al., 2006; Hindré et al., 2008; Pécastaings et al., 2010; Hochstrasser et al., 2019). Legionella pneumophila biofilm formation is not well understood, and only a few bacterial determinants affecting biofilm formation have been described (Abu Khweek and Amer, 2018). The Tat transporter (De Buck et al., 2005), the Legionella collagen‐like (Lcl) adhesin (Duncan et al., 2011; Mallegol et al., 2012) and to a limited extent the type IV pili (Lucas et al., 2006) have been found to be involved in biofilm development.
The flagellum assists biofilm development of various bacteria (Davey and O'Toole, 2000; Petrova and Sauer, 2012). The L. pneumophila flagellum filament protein (flagellin) is encoded by the flaA gene, and its transcription is controlled by the alternative sigma factor FliA (Albert‐Weissenberger et al., 2010). The production of sessile L. pneumophila biomass is regulated by FliA and seems to be independent of FlaA (Mampel et al., 2006; Stewart et al., 2012), but it remains unclear to which extent FlaA mediates the L. pneumophila biofilm architecture.
The pleiotropic transcription factor LvbR (Legionella virulence and biofilm regulator) controls L. pneumophila virulence and biofilm architecture, and its transcription is negatively regulated by the Legionella quorum sensing (Lqs) kinase LqsS (Hochstrasser et al., 2019; Hochstrasser and Hilbi, 2020) (Fig. 1). LvbR negatively regulates the nitric oxide (NO) sensor and di‐guanylate cyclase inhibitor Hnox1, and thus, positively regulates the production of the second messenger cyclic di‐guanosine monophosphate (c‐di‐GMP). Several c‐di‐GMP metabolizing enzymes are implicated in the regulation of L. pneumophila biofilm formation (Carlson et al., 2010; Pécastaings et al., 2016).
Fig. 1.

The L. pneumophila Lqs‐LvbR regulatory network. The Lqs (Legionella quorum sensing) system produces, detects and responds to the small signalling molecule LAI‐1 (Legionella autoinducer‐1, 3‐hydroxypentadecane‐4‐one). The system comprises the autoinducer synthase LqsA, the cognate membrane‐bound sensor kinases LqsS and LqsT, and the prototypic response regulator LqsR. LqsS negatively regulates the expression of lvbR, encoding the transcription factor LvbR (Legionella virulence and biofilm regulator). In turn, LvbR negatively regulates the expression of hnox1, encoding the NO sensor Hnox1, to control the diguanylate cyclase Lpg1057, c‐di‐GMP levels and biofilm architecture. The Lqs‐LvbR network regulates bacterial growth phase switch and motility (flagellin, FlaA), amoeba migration in biofilms and bacterial cluster formation on amoebae, virulence, competence and extracellular filaments, as well as the expression of genes on a 133 kb genomic island, including lvbR.
The Lqs system comprises the autoinducer synthase LqsA (Spirig et al., 2008), the sensor histidine kinases LqsS (Tiaden et al., 2010b) and LqsT (Kessler et al., 2013), and the response regulator LqsR (Tiaden et al., 2007; Tiaden et al., 2008), which dimerizes upon phosphorylation (Schell et al., 2014; Hochstrasser et al., 2020). The Lqs system produces, detects and responds to the signalling molecule LAI‐1 (Legionella autoinducer‐1, 3‐hydroxypentadecane‐4‐one) (Tiaden et al., 2010a; Tiaden and Hilbi, 2012). The system regulates various traits in L. pneumophila (Fig. 1), such as motility and production of flagellin (Schell et al., 2016), virulence (Hochstrasser and Hilbi, 2017; Personnic et al., 2018), the bacterial growth phase switch (Tiaden et al., 2007; Hochstrasser and Hilbi, 2022), expression of a genomic ‘fitness island’ and natural competence for DNA uptake (Hochstrasser and Hilbi, 2017; Personnic et al., 2018), as well as host cell motility (Simon et al., 2015). However, it is not known to what extent the Lqs system controls biofilm formation or architecture.
The interactions between biofilm/sessile bacterial pathogens and amoebae represent crucial ecological processes, which are not well understood on a molecular level. It is currently not known to which extent bacterial factors, such as regulatory systems, virulence factors, the flagellum, or the architecture of biofilms determine the encounter with amoebae. In this study, we visualized and quantified by confocal microscopy the interactions of amoebae with genetically distinct L. pneumophila biofilms. We found that the Lqs regulator LqsR, the transcription factor LvbR, the flagellum and the Icm/Dot T4SS control the migration of A. castellanii through L. pneumophila biofilms, and – with the exception of the T4SS – also regulate bacterial adherence and cluster formation on the amoeba surface.
Results
Comparative proteomics and differential regulation of flagellum production in L. pneumophila ∆ lvbR or ∆ lqsR biofilms
The L. pneumophila transcription factor LvbR regulates biofilm architecture: biofilms formed by a ∆lvbR mutant are homogenous and ‘mat‐like’, while biofilms formed by the parental strain JR32 appear aggregated and ‘patchy’ respectively (Hochstrasser et al., 2019). To further identify components relevant for L. pneumophila biofilm formation, we compared the proteomes of biofilms formed by JR32, ∆lvbR or ∆lqsR mutant strains. To this end, the strains were left to form biofilms in AYE medium for 6 days, bacteria‐associated and secreted proteins were collected, and the proteomes of the mutant strains were compared to the parental strain JR32 (Fig. 2A, Fig. S1). In biofilms formed by the ΔlvbR strain, there were ca. 1.7 times more proteins produced than that in biofilms formed by the ΔlqsR strain (Fig. 2A). Among a total of 1241 proteins identified, 662 or 237 were uniquely produced in ∆lvbR or ∆lqsR biofilms respectively, while 342 were produced in concert in biofilms from both strains, the abundance of which changed synergistically (326 proteins) or reciprocally (16 proteins). Hence, in biofilms formed by the ∆lvbR or ∆lqsR mutants significantly more protein abundances changed jointly rather than reciprocally (Fig. 2A, Fig. S1). These results suggest that the regulators LvbR and LqsR share many common targets in biofilms.
Fig. 2.

Comparative proteomics of L. pneumophila JR32, ∆lvbR and ∆lqsR biofilms and validation of differential P flaA expression.
A. Proteomics was performed with biofilms formed by ΔlvbR or ΔlqsR mutant strains and compared to the biofilms formed by the parental strain JR32. In the Venn diagrams all proteins are shown that are depleted or enriched in mutant biofilms (for details see Materials and Methods). Among the total of 1241 proteins identified, 342 were produced in concert depending on the presence of LvbR and LqsR (overlapping area; brackets: number of LvbR/LqsR‐dependent proteins, which accumulate in parallel (326, black) or reciprocally (16, grey).
B, C. P flaA ‐gfp expression in ∆lvbR and ∆lqsR mutant strains. Legionella pneumophila JR32, ∆lvbR or ∆lqsR strains harbouring the promoter‐reporter constructs P flaA ‐gfp (pCM009) or P flaA ‐gfp and P lvbR ‐lvbR (pRH022) were grown at (B) 25°C or (C) 37°C in AYE medium within microplates while orbitally shaking. GFP fluorescence (relative fluorescence units, RFU) at a gain of 50 and optical density at 600 nm (OD600) were measured over time using a microplate reader. The kinetics of the GFP fluorescence/OD600 values are shown. The data are means and standard deviations of a technical triplicate and representative of two (B) or three (C) independent measurements.
A number of proteins with substantially changed abundances were identified in biofilms formed by the ∆lvbR or ∆lqsR mutant strains (Fig. S1). Specifically, proteins produced by the 133 kb genomic fitness island region I (lpg0973‐lpg1005; e.g. Lpg0987, Lpg0995) and region II (lpg1006‐lpg1096; e.g. Lpg1055) were less (region I) or more (region II) abundant in biofilms formed by the mutant strains (Fig. S1, box 1–2). Moreover, the Legionella collagen‐like (Lcl) adhesin (Lpg2644; SclB tail fibre protein) (Duncan et al., 2011; Mallegol et al., 2012) was found in lower amounts in the ∆lvbR mutants compared to the wild‐type strain (Fig. S1). Components of the Lvh T4SS (LvhB5/Lpg1253, LvhB10/Lpg1247, LvrE/Lpg1244) (Segal et al., 1999) (Fig. S1, box 3) and Icm/Dot‐translocated effector proteins, such as the astacin protease LegP (Lpg2999) (de Felipe et al., 2008), the deAMPylase SidD (Lpg2465) (Neunuebel et al., 2011; Tan and Luo, 2011), the ubiquitin ligase SdeA (Lpg2157) (Qiu et al., 2016), the Rab1 GEF RalF (Lpg1950) (Nagai et al., 2002) and the protein kinase Lpg2603 (Sreelatha et al., 2020) were found in lower amounts in the ∆lvbR and/or ∆lqsR mutants compared to the parental strain JR32 (Fig. S1). In contrast, enzymes implicated in myo‐inositol catabolism, such as IolG (Lpg1652), IolCB (Lpg1651), IolD (Lpg1650) and IolE (Lpg1649) (Manske et al., 2016) were more abundant in biofilms formed by the ∆lvbR or ∆lqsR mutant strains (Fig. S1, box 4), indicating that inositol catabolism in L. pneumophila biofilms might be negatively regulated by LvbR and LqsR.
Intriguingly, in biofilms formed by the ∆lvbR or ∆lqsR mutant strains components of the flagellum were differentially produced, in particular the major flagellar subunit flagellin (FlaA/FliC, Lpg1340) was depleted, while the flagellar hook‐associated protein FliD (Lpg1338), some Flg proteins (Lpg1218‐Lpg1222), the flagellar motor switch protein FliN (Lpg1791) and other Fli proteins (Lpg1758, Lpg1762), as well as the flagellar biosynthesis sigma factor FliA (Lpg1782) were enriched (Fig. S1, box 5–8).
In agreement with these findings, in the ∆lvbR and ∆lqsR mutant strains the expression of a P flaA ‐gfp reporter construct lagged behind and was downregulated at 25°C (Fig. 2B) as well as at 37°C (Fig. 2C), confirming previous results for ∆lqsR (Schell et al., 2016). Complementation of the ∆lvbR mutant phenotype was achieved with a plasmid producing LvbR under control of its natural promoter (P lvbR ‐lvbR) in addition to GFP under control of P flaA (Fig. 2B and C). In agreement with the finding that components of the flagellum are downregulated in the ∆lvbR and ∆lqsR strains, the mutants showed reduced motility upon growth in AYE medium ([Link], [Link]). Taken together, these results suggest that LqsR and LvbR control the production of the flagellum, and in particular, act as positive regulators of FlaA production and flaA transcription.
Based on the finding that the flagellum is regulated by LqsR and LvbR in L. pneumophila biofilms, we assessed whether FlaA is a determinant of biofilm formation and architecture. To this end, GFP‐producing L. pneumophila JR32 or ∆flaA were left to form biofilms in AYE medium for 1, 2, 3 and 6 days at 25°C without disturbance, and biofilm formation, including architecture and adherence to an abiotic surface, was analysed by confocal microscopy (Fig. 3). Under these conditions, the ∆flaA mutant strain showed the same ‘clustered’ and ‘patchy’ biofilm architecture as the parental JR32 strain. In contrast, the ΔlvbR strain formed a ‘mat‐like’ biofilm, as reported previously (Hochstrasser et al., 2019).
Fig. 3.
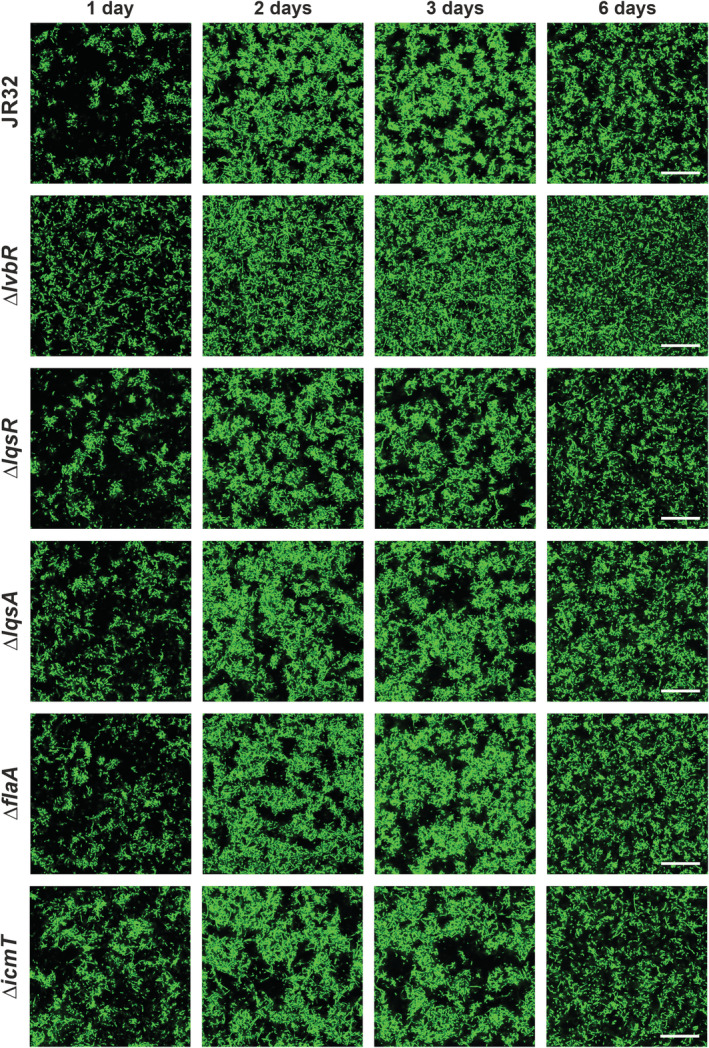
Biofilm formation of L. pneumophila ∆lqs, ∆flaA and ∆icmT mutant strains. Biofilms were initiated with exponential phase L. pneumophila JR32, ∆lvbR, ∆lqsR, ∆lqsA, ∆flaA or ∆icmT mutant strains harbouring pNT28 (constitutive production of GFP) in AYE medium within ibiTreat microscopy dishes. Biofilms were grown at 25°C without mechanical disturbance, and confocal microscopy images of biofilm architecture were acquired at 4 μm above the dish bottom after 1, 2, 3 and 6 days of growth. The images shown are representative of three independent experiments. Scale bars, 30 μm.
Analogously, we assessed the L. pneumophila ΔlqsR, ΔlqsA, ΔlqsS, ΔlqsT and ΔlqsS‐ΔlqsT mutant strains for biofilm formation. The lqs mutant strains formed biofilms with the same morphology as the parental strain JR32 (Fig. 3, Fig. S2A), produced comparable biomass, i.e. the same number of bacteria per ml (Fig. S2B and C), and adhered similarly to the abiotic surface of the microscopy dish (Fig. S3). In summary, biofilms formed by L. pneumophila lacking the major flagellum component FlaA or components of the Lqs system adopt a ‘patchy’ architecture morphologically similar to the parental strain JR32, produce a comparable biomass, and adhere indistinguishably to an abiotic surface. Therefore, under the conditions tested the flagellum is not required to establish L. pneumophila biofilms, and quorum sensing does not control biofilm formation and architecture. However, since the biofilms formed by the ∆lvbR or ∆lqsR mutants regulate flagellum production and differ in their protein pattern compared to JR32 (Fig. S1), a distinct protein composition underlies these morphologically similar biofilms.
LvbR, LqsR and FlaA promote A. castellanii migration through L. pneumophila biofilms
To further investigate the characteristics of genetically different L. pneumophila biofilms, we assessed their interactions with amoebae. The different protein composition profiles of biofilms formed by L. pneumophila JR32 or mutant strains might affect the migration of amoebae through a biofilm. To test this hypothesis, we quantified the migration of A. castellanii through biofilms formed by GFP‐producing L. pneumophila JR32 or the ∆lvbR, ∆lqsR, ∆lqsA or ∆flaA mutant strains (Fig. 4). The biofilms were grown in AYE medium for 6 days at 25°C, and after adding the amoebae, the migration of single cells through the biofilms was tracked by confocal microscopy in the bright field channel ([Link], [Link]).
Fig. 4.

LvbR, LqsR and FlaA promote A. castellanii migration through L. pneumophila biofilms. Biofilms of L. pneumophila JR32, ∆lvbR, ∆lqsR, ∆lqsA or ∆flaA mutant strains harbouring pNT28 (constitutive production of GFP) were grown in AYE medium at 25°C, A. castellanii was added to preformed biofilms after 6 days, and amoeba migration was followed by confocal microscopy.
A. Single amoeba migrating through biofilms were tracked and individually shown as a line in migration plots.
B. The velocities and (C) Euclidean distances of migrating amoebae tracked in (A) were quantified. The data (B, C) are means and standard deviations of the mean derived from all tracked amoebae (n.s., not significant; *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; one‐way ANOVA with Tukey post‐test; JR32 vs. mutant strains). Single amoebae were tracked in three biologically independent experiments with a total of n = 28 amoebae for each bacterial strain.
Under these conditions, the amoebae within JR32 or ΔlqsA biofilms showed a more vigorous migration compared to ΔlvbR, ΔlqsR or ΔflaA biofilms (Fig. 4A). The amoebae velocity was highest in biofilms formed by JR32 (on average ca. 0.22 μm s−1), lower in biofilms generated by ΔlvbR or ΔflaA (ca. 0.15 μm s−1, not significant for ΔlqsA) and lowest in ΔlqsR biofilms (ca. 0.09 μm s−1) (Fig. 4B). The migration distance (Euclidean distance) was ca. 75 μm for amoebae in biofilms formed by JR32, ΔlqsA or ΔflaA and less (but not significantly) for amoebae in biofilms formed by the ΔlvbR or ΔlqsR mutant strains (ca. 40 μm) (Fig. 4C). These observations were rather unexpected, since the JR32 and ΔlqsA strains are more virulent (and thus potentially more toxic) than the ΔlvbR, ΔlqsR or ΔflaA mutant strains. Taken together, A. castellanii amoebae migrate fastest in biofilms established by L. pneumophila JR32, with intermediate speed in ΔlqsA, ΔflaA or ΔlvbR biofilms and slowest in biofilms formed by ΔlqsR. Thus, amoeba migration in L. pneumophila biofilms is positively correlated to the virulence of a strain and promoted by LqsR and LvbR as well as by FlaA.
The Icm/Dot T4SS and the effector LegG1 promote A. castellanii migration through L. pneumophila biofilms
Given the positive correlation between amoeba migration and the L. pneumophila virulence regulators LqsR and LvbR (Fig. 4), we next tested the role of the Icm/Dot T4SS and secreted effector proteins for amoeba migration in L. pneumophila biofilms. To this end, we used the ∆icmT mutant strain, which lacks a functional Icm/Dot T4SS (Böck et al., 2021), is defective for intracellular growth (Segal and Shuman, 1998), and in contrast to the wild‐type strain JR32, does not inhibit cell migration upon infection (Simon et al., 2014). Moreover, we used the ∆legG1 (lpg1976), ∆ppgA (lpg2224) or ∆legG1‐∆ppgA mutant strains, lacking one or two RCC1 repeat effectors, which promote microtubule stabilization, LCV motility and host cell migration (Rothmeier et al., 2013; Swart et al., 2020b). Biofilms formed by the ∆icmT mutant strain in AYE medium for 1, 2, 3 and 6 days at 25°C without disturbance were morphologically similar to the parental strain JR32 (Fig. 3), and the mutant adhered similarly (but due to faster growth more densely) to an abiotic surface (Fig. S3). Therefore, a functional Icm/Dot T4SS is not required for normal L. pneumophila biofilm formation.
We compared the migration of A. castellanii through biofilms formed by L. pneumophila JR32, or the ∆icmT, ∆legG1, ∆ppgA or ∆legG1‐∆ppgA mutant strains (Fig. 5). To this end, the biofilms were grown in AYE medium for 6 days at 25°C, and after adding the amoebae, the migration of single cells through the biofilms was tracked by confocal microscopy in the bright field channel (Movies S4 and S9). In JR32 biofilms the amoebae showed a more vigorous migration compared to biofilms established either by the ∆icmT, ∆legG1, ∆ppgA or the ∆legG1‐∆ppgA double mutant strain (Fig. 5A). The amoeba velocity was highest in biofilms formed by JR32 (on average ca. 0.3 μm s−1), followed by ∆ppgA (ca. 0.28 μm s−1), and significantly lower in ∆legG1 or ∆legG1‐∆ppgA biofilms (ca. 0.15–0.16 μm s−1). Interestingly, amoeba movement was most strongly impaired in ∆icmT biofilms (ca. 0.09 μm s−1) (Fig. 5B). The migration distance (Euclidean distance) was highest for amoebae in biofilms formed by JR32 (ca. 90 μm), lowest in biofilms formed by ΔicmT (ca. 40 μm) and intermediate for amoebae in biofilms formed by the ∆legG1, ∆ppgA or ∆legG1‐∆ppgA mutant strains (ca. 60–70 μm) (Fig. 5C).
Fig. 5.

The Icm/Dot T4SS and the effector LegG1 promote A. castellanii migration through L. pneumophila biofilms. Biofilms of L. pneumophila JR32, ∆icmT, ∆ppgA, ∆legG1 or ∆ppgA‐∆legG1 mutant strains harbouring pNT28 (constitutive production of GFP) were grown in AYE medium at 25°C, A. castellanii was added to preformed biofilms after 6 days and amoeba migration was followed by confocal microscopy.
A. Single amoeba migrating through biofilms were tracked and individually shown as a line in migration plots.
B. The velocities and (C) Euclidean distances of migrating amoebae tracked in (A) were quantified. The data (B, C) are means and standard deviations of the mean derived from all tracked amoebae (n.s., not significant; *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; one‐way ANOVA with Tukey post‐test; JR32 vs. mutant strains). Single amoebae were tracked in three biologically independent experiments with a total of n = 30 amoebae for each bacterial strain.
In summary, compared to biofilms formed by the parental strain JR32, A. castellanii amoebae migrate less vigorously in biofilms formed by mutant strains lacking a functional Icm/Dot T4SS or RCC1 repeat effectors. Amoeba movement was most strongly impaired in ∆icmT biofilms, suggesting that in addition to the RCC1 repeat effectors other Icm/Dot substrates might also contribute to promoting protozoan motility. Hence, amoeba migration in L. pneumophila biofilms is positively correlated to the Icm/Dot‐dependent virulence, similar to what was observed for biofilms formed by regulatory (and flaA) mutant strains (Fig. 4).
LvbR, LqsR and FlaA determine L. pneumophila cluster formation on A. castellanii
To further investigate the processes underlying the migration of A. castellanii through biofilms, we analysed by confocal microscopy the features of different L. pneumophila strains interacting with the amoebae. To this end, GFP‐producing L. pneumophila JR32 or the ∆lvbR, ∆lqsR, ∆lqsA, ∆flaA or ∆icmT mutant strains were left to form biofilms in AYE medium for 6 days at 25°C, and bacteria–amoeba interactions were assessed by confocal microscopy in the GFP or bright field channel respectively (Fig. 6).
Fig. 6.
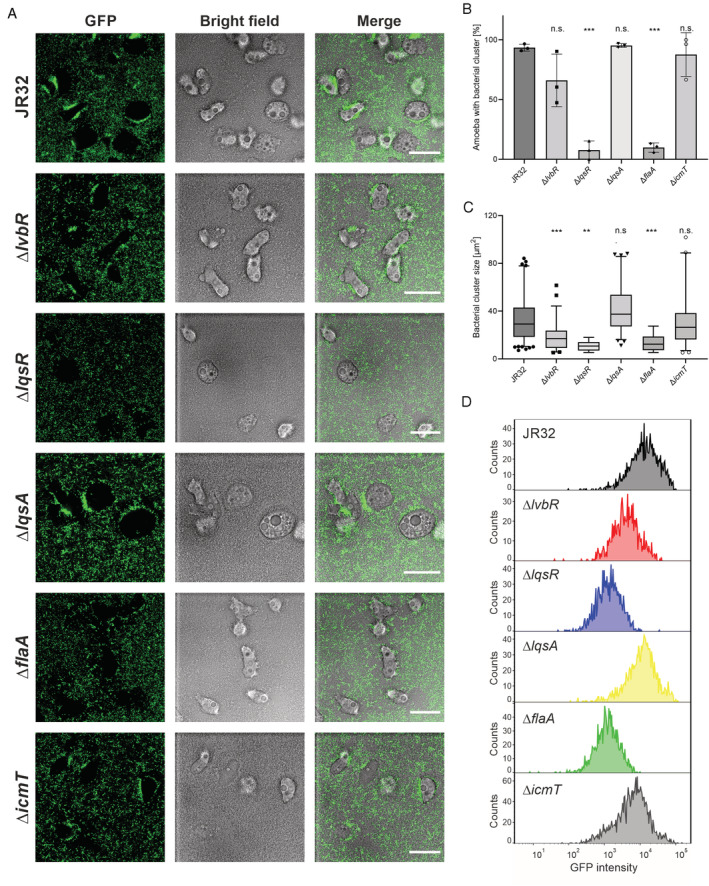
LvbR, LqsR and FlaA determine L. pneumophila cluster formation on A. castellanii. Biofilms of L. pneumophila JR32, ∆lvbR, ∆lqsR, ∆lqsA, ∆flaA or ∆icmT mutant strains harbouring pNT28 (constitutive production of GFP) were grown for 6 days in AYE medium at 25°C, and A. castellanii was added to preformed biofilms.
A. Bacterial adherence and cluster formation were monitored in biofilms containing amoebae by confocal microscopy above the dish bottom. The images shown are representative of three independent experiments. Scale bars, 30 μm.
B. Percentage of amoebae with bacterial cluster (or cluster‐positive amoebae) in the total amoebae population. The columns display the mean value of three biological replicates (in dots) with standard deviations.
C. Quantification by fluorescence microscopy of cluster size on cluster‐positive amoebae. The box‐and‐whisker plots display 5th to 95th percentiles (whiskers), median and quartiles (box) of the pooled results from three biologically independent experiments with a total analysed number of amoebae n = 124 (JR32), n = 60 (∆lvbR), n = 9 (∆lqsR), n = 66 (∆lqsA), n = 8 (∆flaA), or n = 48 (∆icmT). For (B) and (C): n.s., not significant; *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; one‐way ANOVA with Tukey post‐test; JR32 vs. mutant strains.
D. Quantification by flow cytometry (counts vs. fluorescence intensity) of amoeba‐associated, GFP‐positive L. pneumophila (JR32, ∆lvbR, ∆lqsR, ∆lqsA, ∆flaA or ∆icmT harbouring pNT28) using FlowJo software (protocol #1, see Materials and Methods).
Strikingly, amoebae placed onto biofilms generated by JR32, ∆lqsA or ∆icmT were decorated with bacteria that formed large aggregates or clusters adhering to the amoebae (Fig. 6A). Close‐up inspection (Fig. S4A) and 3D‐rendering (Fig. S4B) of these bacterial aggregates revealed multi‐layered clusters decorating the polar or lateral side of amoebae, and preferentially the lagging end of moving amoebae (Movies S4 and S7). Furthermore, ‘trails’ of migrating amoebae were apparent in the surface‐attached layer of JR32 biofilms, suggesting that the migrating amoebae ‘grazed’ through biofilms leaving behind a track devoid of bacteria (Fig. S4C, Movies S4 and S7).
The quantitative assessment of the clusters by confocal microscopy revealed that almost 100% of the amoebae in biofilms formed by the JR32, ∆lqsA, or ∆icmT strains were decorated by bacterial clusters (Fig. 6B). In contrast, ca. 75% of the amoebae were decorated with bacterial clusters in biofilms formed by the ∆lvbR strain, and only ca. 10% of the amoebae were decorated with bacterial clusters in biofilms formed by the ∆lqsR or ∆flaA mutant strains. Moreover, the size of the bacterial clusters was similar for strain JR32, ∆lqsA or ∆icmT (ca. 30 μm2), and significantly smaller for the ∆lvbR, ∆lqsR or ∆flaA mutant strains (ca. 10–20 μm2) (Fig. 6C). The quantification of the clusters by flow cytometry indicated very similar results: the amoeba‐associated fluorescence intensity of GFP‐producing L. pneumophila was highest for the JR32, ∆lqsA, or ∆icmT strains, followed by ∆lvbR, and lowest for the ∆lqsR or ∆flaA mutant strains (Fig. 6D, Fig. S5AB). In summary, these observations indicate that L. pneumophila forms large aggregates or clusters adhering to amoebae within biofilms, and neither the LqsA‐produced quorum‐sensing signalling molecule LAI‐1 nor the Icm/Dot T4SS are required for cluster formation.
In contrast to the bacterial clusters on amoebae migrating in biofilms generated by JR32, ∆lqsA, or ∆icmT, these clusters were barely present and much smaller on amoebae in biofilms formed by ∆lvbR and entirely absent in biofilms formed by ∆lqsR or ∆flaA (Fig. 6). The bacterial clusters on amoebae in ∆flaA biofilms were partially restored by the expression of plasmid‐borne flaA in the mutant strain (Fig. S4D and E). Finally, the mCherry‐producing parental strain JR32 clustered to A. castellanii also in presence of and upon competition with GFP‐producing mutant strains (Fig. S6), indicating that the cluster phenotype is dominant. In summary, these results indicate that the regulatory proteins LvbR and LqsR as well as flagellin (FlaA) not only promote the migration of amoebae in L. pneumophila biofilms but are also implicated in the formation of bacterial clusters on the migrating amoebae, while the autoinducer synthase LqsA and the Icm/Dot T4SS are dispensable for the phenotype.
Amoeba‐adherent L. pneumophila clusters express motility and virulence genes
To further characterize the features of the L. pneumophila clusters on A. castellanii migrating in biofilms, we analysed the activity of promoters serving as prototypic proxies for motility (P flaA ), virulence (P sidC , P ralF ), stress response (P 6SRNA ) and replication (P csrA ). To this end, we constructed single GFP reporters, which produced the fluorescent protein under control of these promoters in AYE broth (Fig. S7A–E). The parental strain JR32 was transformed with transcriptional gfp fusions of the above promoters, and biofilms were grown in AYE medium for 6 days at 25°C. After addition of A. castellanii for ca. 3 h, the promoter activity was analysed by fluorescence microscopy (Fig. 7A). This approach revealed that the P flaA , P sidC and P ralF promoters showed strong activity in the amoeba‐adherent L. pneumophila clusters, while P 6SRNA and P csrA were less or not active. Quantitative assessment of promoter activity by flow cytometry confirmed that P flaA , P sidC and P ralF were strongly induced in amoeba‐associated L. pneumophila, while P 6SRNA and P csrA were less or not active (Fig. 7B and C, Fig. S5C).
Fig. 7.

Amoeba‐adherent L. pneumophila clusters express motility and virulence genes.
A. Biofilms of L. pneumophila JR32 harbouring promoter‐reporter plasmids P flaA ‐gfp (pCM009), P 6SRNA ‐gfp (pRH049), P sidC ‐gfp (pRH035), P ralF ‐gfp (pRH032) or P csrA ‐gfp (pRH031) were grown in AYE medium for 6 days. Acanthamoeba castellanii was added to preformed biofilms and confocal microscopy images of amoebae with adherent bacterial clusters were acquired close to the bottom of microscopy dishes. The images shown are representative of at least two independent experiments. Scale bars, 30 μm. Quantification by flow cytometry of GFP‐positive L. pneumophila JR32 harbouring promoter reporters (P flaA , P 6SRNA , P sidC , P ralF , or P csrA ) using FlowJo software: (B) forward scatter area (FCS‐A) versus fluorescence intensity, or (C) counts versus fluorescence intensity [amoebae‐associated (red) and non‐associated (black)] (protocol #2, see Materials and Methods).
We also tested dual GFP reporter constructs of P flaA P sidC , P ralF , P 6SRNA and P csrA , which in parallel also constitutively produce mCherry. Of these constructs, only P flaA and P 6SRNA produced a robust GFP signal, which was observed in L. pneumophila in broth (Fig. S8A and B, data not shown). Similar to the single reporter constructs, the P flaA promoter was induced in the amoeba‐adherent L. pneumophila clusters in biofilms, while P 6SRNA was not (Fig. S8C). Moreover, usage of the P flaA dual reporter construct confirmed that amoeba‐adherent L. pneumophila clusters were most robustly formed in biofilms formed by strain JR32, less in ∆lvbR biofilms and barely in biofilms formed by the ∆lqsR mutant strain (Fig. S8D). Taken together, these findings are in agreement with the notion that the bacteria in the clusters are in stationary growth phase and motile. In summary, the individual bacteria in the L. pneumophila clusters on A. castellanii migrating in biofilms express the P flaA , P sidC and P ralF promoters, and therefore, are likely in the non‐growing, transmissive and virulent phase.
Discussion
In this study, we investigated the role of the L. pneumophila genotype on biofilm formation and morphology, the migration of amoebae within biofilms and the occurrence of amoeba‐associated bacterial clusters (Table 1). We found that the transcription factor LvbR, the quorum‐sensing response regulator LqsR, the bacterial flagellum (FlaA) and the Icm/Dot T4SS regulate the speed and (except for FlaA) the migration distance of A. castellanii moving through L. pneumophila biofilms (Figs 4 and 5). Furthermore, LvbR, LqsR and FlaA also regulate the adherence and cluster formation of L. pneumophila on the amoeba surface, and the clusters comprise P flaA ‐ and P sidC ‐positive, i.e. motile and virulent bacteria (Figs 6 and 7).
Table 1.
Summary of biofilm‐amoeba interaction and virulence phenotypes.
| Phenotypes | JR32 | ΔlvbR | ΔlqsR | ΔlqsA | ΔflaA | ΔicmT |
|---|---|---|---|---|---|---|
| Biofilm architecture a | p | m | p | p | p | p |
| Amoeba migration | ++ | + | + | ++ | + | + |
| Cluster formation | ++ | + | − | ++ | − | ++ |
| Bacterial virulence | ++ | + | + | ++ | + | − |
Abbreviations: p, ‘patchy’; m, ‘mat’‐like.
Intriguingly, the migration of amoebae through L. pneumophila biofilms was positively correlated to the virulence of the bacterial strains (Table 1). Acanthamoeba castellanii moved slower and less vigorously through biofilms formed by the ∆lvbR and ∆lqsR regulatory mutants (Fig. 4), which are impaired for virulence (Tiaden et al., 2007; Hochstrasser et al., 2019), and also through biofilms formed by the avirulent ∆icmT T4SS mutant strain, or biofilms formed by strains lacking the Icm/Dot‐translocated effectors LegG1 or LegG1 and PpgA (Fig. 5). LegG1 and PpgA are RCC1 repeat effectors, which localize to LCVs or the plasma membrane, target the activation cycle of the small GTPase Ran and activate Ran either at the LCV or the plasma membrane respectively (Rothmeier et al., 2013; Swart et al., 2020b). The activation of Ran GTPase by the L. pneumophila RCC1 repeat effectors promotes microtubule stabilization, LCV motility and cell migration (Rothmeier et al., 2013; Simon et al., 2014; Swart et al., 2020a). Given the cell migration phenotypes of the ∆legG1, ∆ppgA and ∆legG1‐∆ppgA mutant strains, presumably the effectors also positively regulate the microtubule cytoskeleton and cell migration of A. castellanii moving through biofilms.
The amoeba speed was also impaired in biofilms formed by L. pneumophila lacking flaA encoding bacterial flagellin (FlaA), and thus, was positively correlated to the presence of FlaA (Fig. 4). Moreover, the amoebae moved with a lower speed through biofilms formed by the ∆lqsR strain compared to ∆lvbR (Fig. 4B), in agreement with the decreasingly pronounced role of LqsR and LvbR in regulating P flaA (Fig. 2B and C). However, the amoebae moved with an even lower speed through ∆lqsR biofilms compared to ∆flaA. Thus, FlaA is not essential for amoeba migration through L. pneumophila biofilms and LqsR likely regulates other factors implicated in the process.
The mechanism, by which FlaA and the flagellum promote amoeba migration through biofilms is unclear. Perhaps, the amoebae detect and respond to the ‘pathogen‐associated molecular pattern’ flagellin. Overall, the enhanced migration of A. castellanii in biofilms formed by virulent L. pneumophila might be a ‘flight’ reaction. Such a reaction would expand the behavioural repertoire of biofilm‐interacting amoebae, beyond ‘grazing’ on these bacterial communities (Ronn et al., 2002; Huws et al., 2005; Matz and Kjelleberg, 2005).
An obvious explanation for the biofilm genotype‐dependent differences in the migration of amoebae might be distinct biofilm architectures and structures. The homogenous ‘mat’‐like biofilm architecture of ∆lvbR biofilms (Hochstrasser et al., 2019) could cause slower amoeba migration, while amoebae migrate faster through ‘patchy’ biofilms with interspersed clusters formed by wild‐type JR32 bacteria and some mutant strains. However, this seems not to be the case, since the patchy biofilms formed by the ∆lqsR, ∆flaA and ∆icmT mutant strains (Fig. 3) also sustained only slow amoeba migration. Thus, the biofilm morphology and structure do not seem to correlate with the features of amoeba migration. However, we did not label any matrix components, e.g. exopolysaccharides, protein or eDNA, which might account for the differential migration of the amoebae. Hence, the mechanism(s), by which the lack of the regulatory factors (LvbR, LqsR) impede amoeba migration is unclear.
As a general mechanistic concept, LvbR‐ or LqsR‐regulated bacterial factor(s) or feature(s) might trigger and affect the amoeba migration dynamics in L. pneumophila biofilms. Legionella pneumophila strains lacking lvbR (Hochstrasser et al., 2019) or lqsR (Tiaden et al., 2007) are impaired for virulence, and both regulatory factors control the production of Icm/Dot‐translocated effector proteins. In sessile L. pneumophila, some Icm/Dot‐translocated effectors are less abundantly produced by the ∆lvbR or ∆lqsR mutant strains compared to the parental strain (Fig. S1). These include the astacin protease LegP (de Felipe et al., 2008), the deAMPylase SidD (Neunuebel et al., 2011; Tan and Luo, 2011), the ubiquitin ligase SdeA (Qiu et al., 2016), the Rab1 GEF RalF (Nagai et al., 2002) and the protein kinase Lpg2603 (Sreelatha et al., 2020). SdeA and RalF are L. pneumophila effector proteins typically produced in the post exponential growth phase, while LegP and SidD are non‐differentially produced (Aurass et al., 2016). The lack (or lower abundance) of specific effectors might account for the lower migration speed of A. castellanii through biofilms formed by the ∆lvbR or ∆lqsR mutants, analogously to what has been observed for the ∆icmT, ∆legG1, ∆ppgA and ∆legG1‐∆ppgA mutant strains (Fig. 5). Alternatively or additionally, the ∆lvbR or ∆lqsR mutants might be defective for the production of adhesins or components of the extracellular biofilm matrix. As the amoebae were added to L. pneumophila biofilms formed over 6 days, such bacterial structures might indeed play a role in determining amoeba migration.
In previous studies, the inhibition of migration and chemotaxis of D. discoideum, macrophages and neutrophils was positively correlated with the virulence of L. pneumophila (Simon et al., 2014; Hochstrasser et al., 2019). While the parental strain JR32 dose‐dependently inhibited phagocyte migration in under‐agar and transwell assays, the ∆icmT mutant did not. Compared to JR32, the less virulent ∆lvbR mutant strain inhibited D. discoideum migration less efficiently, but more efficiently than ∆icmT (Hochstrasser et al., 2019). Legionella pneumophila ∆legG1 even hyper‐inhibited phagocyte migration, indicating that microtubule stabilization by the RCC1 repeat effector promotes cell motility (Simon et al., 2014). As outlined above, presumably the RCC1 repeat effectors also positively regulate the microtubule cytoskeleton and cell migration of A. castellanii moving through biofilms (Fig. 5).
In the previous migration and chemotaxis studies, the phagocytes were infected with L. pneumophila strains for 1 h prior a 4 h migration/chemotaxis assay, and therefore, had already taken up bacteria (Simon et al., 2014; Hochstrasser et al., 2019). Accordingly, the effect of L. pneumophila on eukaryotic cell migration seems to depend on whether planktonic bacteria are being taken up or cluster extracellularly. In agreement with this notion, the efficient uptake of L. pneumophila by amoebae and macrophages is promoted by LvbR (Hochstrasser et al., 2019), LqsR (Tiaden et al., 2007; Tiaden et al., 2008), the flagellum (FlaA) (Dietrich et al., 2001) and the Icm/Dot T4SS (Hilbi et al., 2001; Weber et al., 2006).
Another intriguing observation was the adherence and cluster formation of L. pneumophila on amoebae migrating in biofilms. Cluster formation was controlled by LvbR, LqsR and FlaA, but not by LqsA or the Icm/Dot T4SS, since the ∆lvbR, ∆lqsR and ∆flaA mutants were defective for cluster formation, while ∆lqsA or ∆icmT formed clusters like the parental strain JR32 (Fig. 6). Legionella pneumophila lacking flaA was severely impaired for cluster formation, and thus, FlaA might act as an adhesin and promote binding to the amoebae. While the bacterial flagellum is known to act as an adhesion factor (Haiko and Westerlund‐Wikstrom, 2013), it is unclear whether the flagellum per se or flagella‐based motility is required for cluster formation of L. pneumophila. LvbR and LqsR regulate the production of the L. pneumophila flagellum filament protein FlaA (Lpg1340) and the transcription of flaA (Fig. S1, Fig. 2B and C), and thus, possibly contribute to cluster formation on amoebae. Finally, in agreement with a role of LqsR for the regulation of bacterial adherence and cluster formation, LqsR (but not LqsA) was previously found to be a regulator of ‘extracellular filaments’, which form a network and prevent the sedimentation of the bacteria (Tiaden et al., 2007).
In addition to the flagellum, other L. pneumophila factors possibly promote bacterial cluster formation on the amoeba surface in the biofilms. LvbR as well as LqsR are positive regulators of Tfp pili (pilus retraction ATPase PilT, Lpg0987) (Fig. S1), which might function as adhesins. Lcl (Lpg2644) is another candidate, since this adhesin mediates the adherence to host cells (Vandersmissen et al., 2010) and is implicated in bacterial adhesion during biofilm formation (Duncan et al., 2011; Mallegol et al., 2012). Intriguingly, Lcl is less abundant in biofilms formed by the ∆lvbR mutant strain (Fig. S1), which might contribute to the reduced adherence and cluster formation of the mutant on amoebae (Fig. 6). In general, bacterial factors contribute individually or in concert to the extracellular biofilm matrix, which might promote adherence and clustering of L. pneumophila on the surface of amoebae.
The function of the L. pneumophila clusters on amoebae in biofilms is unclear. Successful and efficient intracellular replication of L. pneumophila depends on adherence to host cells (Cirillo et al., 2001; Chang et al., 2005; Duncan et al., 2011), and thus, cluster formation might facilitate L. pneumophila uptake by host cells within biofilms. Alternatively or additionally, bacterial adherence to amoebae and subsequent cell migration possibly helps L. pneumophila to disseminate in the environment. A similar clustering phenomenon was recently reported for planktonic Listeria monocytogenes, a food‐borne, extracellular Gram‐positive bacterium. Listeria monocytogenes formed densely packed aggregates on A. castellanii and A. polyphaga called ‘backpacks’, and the formation was dependent on flagella‐based motility (Doyscher et al., 2013). However, L. monocytogenes does not persist in Acanthamoeba spp. and simply serves as food for the amoebae. In contrast, L. pneumophila does not serve as food but rather consumes and destroys the amoebae. Hence, L. pneumophila might subvert a mechanism employed by A. castellanii to trap and feed on bacteria, in order to efficiently infect protozoan host cells.
Comparative proteomics of biofilms formed by JR32, ∆lvbR or ∆lqsR mutant strains revealed that LvbR and LqsR are implicated in the regulation of the flagellum, the Lvh T4SS, Icm/Dot‐translocated effectors and proteins encoded by the 133 kb genomic island regions I and II (Fig. S1). Comparative transcriptomics of sessile and planktonic JR32, ∆lvbR or ∆lqsR mutant strains indicated similar target genes of the regulators, including the 133 kb genomic island region I (lpg0973‐1005) and region II (lpg1006‐1096), the lvh region (lpg1244‐1259) and various genes involved in flagellar biosynthesis (lpg1216‐1226, lpg1337‐1340, lpg1759‐1763, lpg1780‐1789 and lpg1791‐1792) (Hochstrasser et al., 2019). Taken together, these studies provide insights on the components of L. pneumophila biofilms and their interactions with amoebae. Further studies will address the functions and mechanisms underlying the migration of amoebae in L. pneumophila biofilms and bacterial cluster formation on the amoebae.
Experimental procedures
Growth and motility of bacteria, cultivation of amoebae
Legionella pneumophila strains (Table 2) were grown on CYE agar plates for 3 days (Feeley et al., 1979), or were grown to exponential phase in liquid cultures with N‐(2‐acetamido)‐2‐aminoethanesulfonic acid‐buffered yeast extract (AYE) medium (Horwitz, 1983) at 37°C on a wheel (80 rpm) for approximately 18 h. AYE medium was supplemented with chloramphenicol (Cm; 5 μg ml−1) to maintain plasmids if required.
Table 2.
Bacterial strains and plasmids used in this study.
| Strain or plasmid | Relevant properties a | Reference |
|---|---|---|
| E. coli | ||
| TOP10 | Invitrogen | |
| L. pneumophila | ||
| AK01 (ΔlqsT) | JR32 lqsT::Km | Kessler et al. (2013) |
| AK02 (ΔlqsS‐ΔlqsT) | JR32 lqsS::Km lqsT::Gm | Kessler et al. (2013) |
| AK03 (ΔlvbR) | JR32 lvbR::Km | Hochstrasser et al. (2019) |
| ER01 (ΔlegG1) | JR32 legG1::Km | Rothmeier et al. (2013) |
| GS3011 (ΔicmT) | JR32 icmT3011::Km | Segal and Shuman (1998) |
| JR32 | L. pneumophila Philadelphia‐1, serogroup 1, salt‐sensitive isolate of AM511 | Sadosky et al. (1993) |
| LS01 (ΔppgA‐ΔlegG1) | JR32 ppgA::Gm legG1::Km | Swart et al. (2020b) |
| LS03 (ΔppgA) | JR32 ppgA::Km | Swart et al. (2020b) |
| NT02 (ΔlqsA) | JR32 lqsA::Km | Tiaden et al. (2010b) |
| NT03 (ΔlqsR) | JR32 lqsR::Km | Tiaden et al. (2007) |
| NT05 (ΔlqsS) | JR32 lqsS::Km | Tiaden et al. (2010b) |
| ΔflaA | JR32 flaA::Km | Weber et al. (2012) |
| Plasmids | ||
| pAK18 | pMMB207C‐P lvbR ‐lvbR, gfp (constitutive) | Hochstrasser et al. (2019) |
| pCM009 | pMMB207C‐P flaA ‐gfp (ASV) | Schell et al. (2016) |
| pNP102 | pMMB207C, ΔlacI q, mcherry (constitutive) | Steiner et al. (2017) |
| pNT28 | pMMB207C, gfp (constitutive) | Tiaden et al. (2007) |
| pNT31 | pMMB207C‐P lqsS ‐lqsS, gfp (constitutive) | Tiaden et al. (2010b) |
| pRH022 | pMMB207C‐P lvbR ‐lvbR‐P flaA ‐gfp (ASV) | This study |
| pRH031 | pMMB207C‐P csrA ‐gfp (ASV), Cm | This study |
| pRH032 | pMMB207C‐P ralF ‐gfp (ASV), Cm | Striednig et al. (2021) |
| pRH035 | pMMB207C‐P sidC ‐gfp (ASV), Cm | Striednig et al. (2021) |
| pRH046 | pMMB207C‐P csrA ‐gfp (AAV), mcherry (constitutive), Cm | This study |
| pRH047 | pMMB207C‐P ralF ‐gfp (AAV), mcherry (constitutive), Cm | This study |
| pRH049 | pMMB207C‐P 6SRNA ‐gfp (ASV), Cm | This study |
| pRH050 | pMMB207C‐P sidC ‐gfp (AAV), mcherry (constitutive), Cm | This study |
| pSB001 | pMMB207C‐P flaA ‐flaA, gfp (constitutive) | This study |
| pSN7 | pMMB207C‐P flaA ‐gfp (AAV), mcherry (constitutive), Cm | Personnic et al. (2019) |
| pSN25 | pMMB207C‐P 6SRNA ‐gfp (AAV), mcherry (constitutive), Cm | Personnic et al. (2021) |
Abbreviations: Cm, chloramphenicol resistance; Km, kanamycin resistance; Gm, gentamicin resistance.
Bacterial motility was assessed basically as described (Schell et al., 2016). Briefly, L. pneumophila JR32 or the ∆lvbR or ∆lqsR mutant strains were grown for 3 days on CYE/Cm agar plates. Liquid cultures were inoculated in AYE medium at an OD600 of 0.1 and grown for 21 h at 37°C. After 21 h, bacterial cultures were diluted to an OD600 of 0.15, and 700 μl were filled into ibidi‐microscopy dishes. Bacteria were tracked for 90 with 1.298 s intervals using a Leica SP8 confocal microscope (63× oil objective). Bacterial motility was visualized using the ImageJ software. The [Link], [Link] are representative of two independent experiments.
Acanthamoeba castellanii (ATCC 30234, lab collection) was cultured in PYG medium (Moffat and Tompkins, 1992; Segal and Shuman, 1999) at 23°C using proteose from Becton Dickinson Biosciences and yeast extract from Difco.
Molecular cloning
Plasmids used in this study are listed in Table 2. DNA cloning was carried out according to standard protocols, plasmids were isolated using commercially available kits (Qiagen, Macherey‐Nagel), and DNA fragments were amplified using the primers listed in Table S1. All the constructs were verified by DNA sequencing. The complementation plasmids pRH022 (P lvbR ‐lvbR) and pSB001 (P flaA ‐flaA) were generated by amplifying the corresponding gene with its putative natural promoter region using the primer pair oRH040/oRH041 or oRH174/oRH175 and pAK18 or genomic JR32 DNA as template. The NEBuilder HiFi DNA Assembly reaction was used to clone the PCR product into BspQI‐digested pCM009 (Schell et al., 2016) resulting in pRH022 or into BamHI‐digested pNT31 (Tiaden et al., 2010b) thereby replacing P lqsS ‐lqsS and resulting in pSB001.
Single reporter constructs pRH031, and pRH049 containing a transcriptional fusion P csrA ‐gfp(ASV), or P 6SRNA ‐gfp(ASV) were generated by PCR amplification of the putative promoter regions from genomic JR32 DNA using the primer pair oRH128/oRH129, or oRH215/oRH216. The putative promoter regions were defined as the upstream region of the corresponding genes with the following sizes: P csrA (515 bp, lpg0781) and P 6SRNA (534 bp, ssrS, between lpg0877 and lpg0876). The PCR products were cloned into SacI‐ and XbaI‐digested pCM009 (Schell et al., 2016) using the NEBuilder HiFi DNA Assembly reaction, thereby replacing P flaA .
For construction of the dual reporter constructs pRH046, pRH047 and pRH050 harbouring P csrA ‐gfp(AAV), P ralF ‐gfp(AAV) or P sidC ‐gfp(AAV), the putative promoter regions were amplified by PCR with the primer pairs oRH182/oRH210, oRH180/oRH211 or oRH217/oRH218 and pRH031, pRH032 or pRH035 as template. The putative promoter regions were defined as the upstream region of the corresponding genes with the following sizes: P ralF (515 bp, lpg1950) and P sidC (507 bp, lpg2511). The resulting PCR products were inserted into BamHI‐ and BglII‐digested pSN7 (Personnic et al., 2019) using the NEBuilder HiFi DNA Assembly reaction, thereby exchanging P flaA .
Kinetics of GFP reporter constructs
Legionella pneumophila strains (Table 2) harbouring GFP reporter constructs (single reporters: pCM009, pRH022, pRH031, pRH032, pRH035, pRH049, or dual reporters: pRH046, pRH047, pSN7, pSN25) were grown on CYE/Cm agar plates for 3 days, followed by cultivation in AYE/Cm medium at 37°C on a rotating wheel (80 rpm) for approximately 18 h. The strains were diluted in AYE/Cm medium to an initial OD600 of 0.2, and 100 μl was distributed in triplicates into wells of a black clear bottom 96‐well plate (polystyrene, 353219, Falcon). The plate was incubated at 25°C or 37°C while orbitally shaking, and GFP fluorescence (excitation, 485 nm; emission, 528 nm; gain, 50 or 100) as well as OD600 (bacterial growth) were measured over time using a microplate reader (Synergy H1 or Cytation5 Hybrid Multi‐Mode Reader, BioTek). A blank value (AYE medium) was subtracted from all values, and numbers are expressed as relative fluorescence units (RFU), OD600 or RFU/OD600 quotient.
Legionella pneumophila biofilm formation
For analysis of biofilm architecture or Legionella–amoeba interactions, L. pneumophila biofilms were prepared, monitored and analysed as described (Hochstrasser and Hilbi, 2019). Briefly, L. pneumophila strains harbouring pNT28, pNP102 or single/dual reporter constructs (Table 2) were grown for 3 days at 37°C on CYE agar plates supplemented with Cm (5 μg ml−1) and subsequently cultivated in AYE/Cm medium on a rotating wheel (80 rpm) at 37°C for approximately 17–18 h. The exponential phase bacteria were inoculated at an optical density (OD600) of 0.3 in 2 ml AYE/Cm medium placed into ibiTreat microscopy dishes (ibidi), incubated at 25°C for the time indicated while avoiding any mechanical disturbance, and optionally stained with DAPI (1 μg ml−1). The biofilm architecture was monitored by confocal fluorescence microscopy (Leica TCS SP5, 63× oil objective) by acquiring images at the dish bottom (0 μm, attachment layer) or at a level of 4 μm above the dish bottom. For competition assays, biofilms were generated analogously with an inoculation OD600 of 0.15 for each strain (start ratio of 1:1) using L. pneumophila JR32 harbouring pNP102 (mCherry) and mutant strains harbouring pNT28 (GFP).
To quantify biofilm formation, L. pneumophila strains harbouring pNT28 were left to form biofilms at 25°C for 6 days as described above. The biofilms were re‐suspended by excessive pipetting until the bottom of the dish appeared clear. The suspension was transferred into a falcon tube, excessively vortexed (to avoid that the bacteria stick together), and the OD600 was determined (usually 3.3–4.1). Subsequently, the suspension was diluted 1:100 (beads: bacteria) with a microsphere standards suspension (Bacteria counting kit, Invitrogen). The biofilm mass (bacteria ml−1) was quantified using a Fortessa II flow cytometer and Diva software. Bacteria and microsphere standard were determined by employing a forward (FSC, 650 V) and sideward scatter (SSC, 230 V), with a threshold of 200 each. Additionally, GFP production (Blue 530_30, 350 V) was examined to identify L. pneumophila. 100000 events of each resuspended biofilm were recorded. Immediately before measuring, samples were strongly vortexed for 8 s. The data were analysed with the software FlowJo. The gating protocol was as follows: The beads were identified using a control sample (AYE/Cm without bacteria) that was supplemented with beads as a reference (gate 1, FSC vs. SSC). The number of events counted in the microsphere gate provides an accurate estimate of the volume analysed. The L. pneumophila populations were determined in pseudocolour graphs (gate 2, GFP vs. FSC) using a resuspended biofilm sample (of parental strain JR32) without beads as a reference. Bacterial density in the biofilm was determined from the ratio of L. pneumophila (gate 2) to beads (gate 1).
Legionella–amoeba interactions
For confocal fluorescence microscopy, A. castellanii amoebae were detached after 2–3 days of growth and diluted in fresh PYG medium to a density of 1.5 × 105 cells ml−1. 1 ml of amoebae suspension was carefully added to preformed L. pneumophila biofilms grown for 6 days (as described above) resulting in a final amoebae concentration of 5 × 104 cells ml−1 in the dish. The dish was incubated for approximately 3 h (images) or 15 min (time‐lapse stacks) at 25°C to allow the amoebae to settle toward the bottom of the dish. Legionella–amoeba interactions were visualized by confocal fluorescence microscopy (Leica TCS SP5, 63× oil objective) by acquiring images or time‐lapse stacks and 3D images or movies were generated using the program ImageJ. For further analysis of amoeba migration, single‐cell tracking was manually performed in the bright field channel using time‐lapse stacks and the program ImageJ. Time series of ca. 10 amoebae localizing above the dish bottom were recorded for the same time (25 or 30 min). Only viable, irregularly shaped amoebae were considered (rounded and clustered amoebae were ignored, except in the case of the ΔicmT mutant strain, which caused a larger portion of the amoebae to round up). The ibidi program ‘Chemotaxis and Migration Tool’ was used to generate migration plots and to quantify the amoeba velocity (Hochstrasser and Hilbi, 2019).
The total number of amoebae and the number of amoebae with a bacterial cluster (or cluster‐positive amoebae) were counted manually from the microscopy images of L. pneumophila strains harbouring pNT28 (constitutive production of GFP). The percentage of cluster‐positive amoebae was assessed for each of three independent biological experiments. Subsequently, for each cluster‐positive amoebae, the cluster size was quantified with ImageJ (Version 1.53f51). In brief, a detection mask was created for each microscopy image with its GFP channel (‘Image’ > ‘Adjust’ > ‘Threshold: 100–255’). Then the size of all clusters in the image was analysed using ‘Analyse particle’ function (‘Analyse’ > ‘Analyse particle’ > ‘Size 10 micron – Infinity’). The size of clusters was quantified for L. pneumophila strains harbouring pNT28 (constitutive production of GFP) in three independent biological experiments.
For flow cytometry, A. castellanii amoebae were added to preformed biofilms grown for 6 days at 25°C as described above, optionally stained with DAPI (1 μg ml−1) and incubated for approximately 2–3 h at 25°C. Biofilms were harvested by resuspending the bacteria and amoebae, followed by centrifugation for 10 min at 100 g (biofilms including amoebae), or for 5 min at 4400 rpm (control biofilms without amoebae). Samples were fixed with 300 μl 4% PFA in PBS for approximately 30 min at RT, washed once in 300 μl PBS and resuspended in 150 μl PBS for flow cytometry analysis.
The size of GFP‐positive L. pneumophila clusters on A. castellanii in bacterial biofilms was quantified by flow cytometry as amoeba‐associated fluorescence using a Fortessa II flow cytometer (BD LSR II Fortessa) and Diva software. Different populations were identified employing a forward (FSC, 430 V) and sideward scatter (SSC, 270 V) gating, with a threshold of 200 each, and 10000 events per sample were recorded. Furthermore, the populations were examined for GFP production [Blue 530_30, 350 V (gating protocol #1) or 400 V (gating protocol #2)]. The data were analysed with the software FlowJo. The gating protocol #1 for strains containing plasmid pNT28 was as follows: amoebae populations were identified using an amoeba only sample as a reference (gate 1). Cluster formation was visualized by a GFP‐shift of the amoebae population visible in pseudocolour graphs (including the L. pneumophila population) and histograms (not including the L. pneumophila population, gate 1). The gating protocol #2 for strains containing promoter expression constructs was as follows: amoebae/L. pneumophila populations were identified using amoebae/L. pneumophila only samples as a reference (gates 1 and 2). The GFP signal of the two populations was visualized together in a histogram (gate 1 and 2) and in pseudocolour graphs (no gating).
Comparative proteomics
Biofilms of L. pneumophila JR32, ΔlvbR and ΔlqsR were initiated as described above and incubated for 6 days at 25°C. The biofilms were harvested by resuspending the bacteria, followed by centrifugation (5 min, 12850 g, 4°C) and resulting supernatants (designated ‘s’, extracellular) and cell pellets (designated ‘p’, intracellular) were analysed separately.
For extracellular sample preparation, proteins in 1 ml of supernatant were enriched using StrataClean beads and eluted by the use of a 1D‐SDS gel according to the established protocol for GeLC‐MS samples (Bonn et al., 2014). Cell pellets (intracellular samples) were resuspended in TE‐buffer pH 7.4 (50 mM Tris, 10 mM EDTA) and disrupted by bead beating. After centrifugation the protein concentration was determined using the Bradford assay and 20 μg protein was in solution digested (Maass et al., 2011) and purified using C18‐Stage tips (Thermo Fisher Scientific). Extracellular proteins (with three biological replicates for every sample) and intracellular proteins (with three biological replicates and three technical replicates each for every sample) were analysed with a QExactive‐Mass spectrometer. Peptide samples were injected with the same volume to allow label‐free relative protein quantification between biological samples.
To process the data, *.raw files were searched against the database using MaxQuant (1.6.0.16). The database for L. pneumophila Philadelphia‐1 was downloaded from Uniprot on March 17th, 2017 and supplemented with the LvbR sequence and common laboratory contaminations resulting in 2973 entries. The results were filtered using 1% FDR in MaxQuant and at least two unique peptides were necessary for protein identification (filtered in Perseus vs. 1.5.3.0). For quantification the protein had to be detected in at least two replicates of one sample. Log2‐transformed LFQ‐values as proxy for protein abundance were exported and used for statistical analysis in TMEV. Only proteins with a minimal log2‐fold change of −0.8 or 0.8 compared to JR32 were considered. In cases, where proteins were detected in ΔlvbR or ΔlqsR but not in JR32 biofilms (‘on’), or where proteins were detected in JR32 but not in ΔlvbR or ΔlqsR biofilms (‘off’), the maximum values of 27.4 and 2−5 were used as ratios and coloured dark blue or dark red in the heat map (Fig. S1) respectively.
Supporting information
Fig. S1. Heat map of proteins depleted or enriched in ∆lvbR and ∆lqsR biofilms compared to JR32.
Comparative proteomics was performed with biofilms formed by L. pneumophila JR32, ΔlvbR or ΔlqsR. In the heat map all proteins are shown that are depleted or enriched in mutant biofilms (for details see Materials and Methods). Biofilm bacteria were harvested by centrifugation and supernatants (designated ‘s’, extracellular) and cell pellets (designated ‘p’, intracellular) were analysed separately. The proteins are listed by ascending Lpg numbers. The colour code indicates the level of regulation: the darker the colour the stronger the corresponding gene is regulated (red: depleted proteins, max. 2−5; blue: accumulated proteins, max. 27.4). Boxes highlight regions of interest, which are further discussed. ‘NaN’: not a number.
Fig. S2. Biofilm formation and quantification of L. pneumophila strains.
(A) Biofilms were initiated with exponential phase L. pneumophila ∆lqsS, ∆lqsT or ∆lqsS‐∆lqsT mutant strains harbouring pNT28 (constitutive production of GFP) in AYE medium within ibiTreat microscopy dishes. Biofilms were grown at 25°C without mechanical disturbance and confocal microscopy images of biofilm architecture were acquired at 4 μm above dish bottom after 1, 2, 3 and 6 days of growth. The images shown are representative of at least two independent experiments. Scale bars, 30 μm. (B, C) Quantification of L. pneumophila biofilms grown for 6 days. (B) Pseudocolour graph (GFP intensity vs. FSC) depicting the gate for GFP‐producing L. pneumophila (identified by fluorescence) and the beads population (identified by SSC vs. FSC). (C) Means and standard deviation (bacteria/ml) of biological triplicates from 100′000 events of each biofilm suspension (differences between wild‐type and mutant L. pneumophila are statistically not significant).
Fig. S3. Surface adherence of biofilms formed by L. pneumophila JR32 and mutant strains.
Biofilms were initiated with exponential phase L. pneumophila JR32, ∆lvbR, ∆lqsR, ∆lqsA, ∆lqsS, ∆lqsT, ∆lqsS‐∆lqsT, ∆flaA or ∆icmT mutant strains harbouring pNT28 (constitutive production of GFP) in AYE medium within ibiTreat microscopy dishes. Biofilms were grown at 25°C without mechanical disturbance and confocal microcopy images of biofilm attachments were acquired at the dish bottom (0 μm) after 1, 2, 3 and 6 days of growth. The images shown are representative of at least two independent experiments. Scale bars, 10 μm.
Fig. S4. L. pneumophila cluster formation on A. castellanii in JR32 or mutant strain biofilms.
Biofilms of (A) L. pneumophila JR32, ∆lvbR, ∆lqsR, ∆lqsA, ∆flaA or ∆icmT mutant strains harbouring pNT28 (constitutive production of GFP), or (B, C) strain JR32 harbouring pNT28 were grown for 6 days in AYE medium at 25°C, and A. castellanii was added to preformed biofilms. Bacterial adherence and cluster formation were monitored by confocal microscopy (A, B) ca. 4 μm above or (C) at the dish bottom ("clusters": arrow heads, "trails": arrows). (D, E) Biofilms of L. pneumophila JR32 or ∆flaA mutant strains constitutively producing GFP (pNT28) or GFP and FlaA under control of its promoter (∆flaA/+flaA, pSB001) were grown for 6 days in AYE medium at 25°C, and A. castellanii was added to preformed biofilms. Confocal microscopy images of (D) overview and (E) zoom‐in of L. pneumophila adherence and cluster formation on amoebae. The images shown are representative of three independent experiments. Scale bars, 30 μm.
Fig. S5. Quantification of amoeba‐associated L. pneumophila clusters by flow cytometry.
Quantification by flow cytometry (forward scatter area, FSC‐A, vs. fluorescence intensity) using FlowJo software of (A) L. pneumophila strain JR32 or A. castellanii, (B) amoebae‐associated or non‐associated, GFP‐positive L. pneumophila JR32, ∆lvbR, ∆lqsR, ∆lqsA, ∆flaA or ∆icmT harbouring pNT28 (protocol #1, see Material and Methods), or (C) non‐associated, GFP‐positive L. pneumophila JR32 harbouring promoter expression reporters (P flaA , P 6SRNA , P sidC , P ralF , or P csrA ) (protocol #2, see Material and Methods).
Fig. S6. Competition of mCherry‐producing L. pneumophila JR32 and GFP‐producing mutants for cluster formation on amoebae.
Biofilm formation was initiated with a 1:1 ratio of exponential phase mCherry‐producing L. pneumophila JR32 (pNP102) and GFP‐producing JR32 or mutant strains (pNT28). Biofilms were grown in AYE medium within ibiTreat microscopy dishes at 25°C for 6 days. A. castellanii were added to preformed biofilms, and amoebae with adherent bacterial clusters were monitored by confocal microcopy above dish bottom. The images shown are representative of at least two independent experiments. Scale bars, 30 μm.
Fig. S7. Gene expression kinetics of L. pneumophila JR32 in AYE medium using single promoter reporters.
L. pneumophila JR32 harbouring (A) P flaA ‐gfp (pCM009), (B) P 6SRNA ‐gfp (pRH049), (C) P sidC ‐gfp (pRH035), (D) P ralF ‐gfp (pRH032) or (E) P csrA ‐gfp (pRH031) reporter constructs were grown at 37°C in AYE medium in microplates while orbitally shaking. GFP fluorescence (relative fluorescence units, RFU) at a gain of 50 and optical density at 600 nm (OD600) were monitored over time using a microplate reader. The kinetics of the GFP fluorescence or OD600 values are shown and depicted by the left or right y‐axis respectively. The data are means and standard deviations of technical triplicates and representative of three independent measurements.
Fig. S8. Gene expression kinetics of L. pneumophila JR32 in AYE medium and in amoeba‐adherent bacteria in biofilms using dual promoter reporters.
L. pneumophila JR32 harbouring (A) P flaA ‐gfp (pSN7) or (B) P 6SRNA ‐gfp (pSN25) reporter constructs were grown at 37°C in AYE medium within microplates while orbitally shaking. GFP fluorescence (relative fluorescence units, RFU) at a gain of 50 and optical density at 600 nm (OD600) were monitored over time using a microplate reader. The kinetics of the GFP fluorescence (left y‐axis) or OD600 (right y‐axis) values are shown. The data are means and standard deviations of technical triplicates and representative of three independent measurements. (C, D) Biofilms of L. pneumophila JR32, ∆lvbR or ∆lqsR harbouring dual reporter plasmids expressing P tac ‐mcherry (constitutive) and P flaA ‐gfp (pSN7) or P 6SRNA ‐gfp (pSN25) were grown in AYE medium for 6 days. A. castellanii was added to preformed biofilms, and confocal microcopy images of amoebae with adherent bacterial clusters were acquired close to the bottom of microscopy dishes. The images shown are representative of 2–3 independent experiments. Scale bars, 30 μm.
Table S1. Oligonucleotides used in this study.
Movie S1. Motility of L. pneumophila JR32.
Movie S2. Motility of L. pneumophila ∆lvbR.
Movie S3. Motility of L. pneumophila ∆lqsR.
Movie S4. A. castellanii migrating through JR32 biofilm.
Movie S5. A. castellanii migrating through ∆lvbR biofilm.
Movie S6. A. castellanii migrating through ∆lqsR biofilm.
Movie S7. A. castellanii migrating through ∆lqsA biofilm.
Movie S8. A. castellanii migrating through ∆flaA biofilm.
Movie S9. A. castellanii migrating through ∆icmT biofilm.
Acknowledgements
The work in the group of H.H. was supported by the Swiss National Science Foundation (SNF; 31003A_175557, 310030_200706), the Novartis Foundation for Medical‐Biological Research (21C165). The work in the group of D.B. was supported by the Federal Ministry of Education and Research (BMBF; grant 031A410B). Open Access Funding provided by Universitat Zurich.
Data Availability
The MS proteomics data discussed in this publication have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (Vizcaino et al., 2016; Perez‐Riverol et al., 2019) with the dataset identifier PXD030337 (Reviewer account details: Username, reviewer_pxd030337@ebi.ac.uk; Password, a2Q4ygNx).
References
- Abdel‐Nour, M. , Duncan, C. , Low, D.E. , and Guyard, C. (2013) Biofilms: the stronghold of Legionella pneumophila . Int J Mol Sci 14: 21660–21675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu Khweek, A. , and Amer, A.O. (2018) Factors mediating environmental biofilm formation by Legionella pneumophila . Front Cell Infect Microbiol 8: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert‐Weissenberger, C. , Sahr, T. , Sismeiro, O. , Hacker, J. , Heuner, K. , and Buchrieser, C. (2010) Control of flagellar gene regulation in Legionella pneumophila and its relation to growth phase. J Bacteriol 192: 446–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aurass, P. , Gerlach, T. , Becher, D. , Voigt, B. , Karste, S. , Bernhardt, J. , et al. (2016) Life stage‐specific proteomes of Legionella pneumophila reveal a highly differential abundance of virulence‐associated Dot/Icm effectors. Mol Cell Proteomics 15: 177–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigot, R. , Bertaux, J. , Frere, J. , and Berjeaud, J.M. (2013) Intra‐amoeba multiplication induces chemotaxis and biofilm colonization and formation for Legionella . PLoS One 8: e77875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böck, D. , Hüsler, D. , Steiner, B. , Medeiros, J.M. , Welin, A. , Ramomska, K.A. , et al. (2021) The polar Legionella Icm/Dot T4SS establishes distinct contact sites with the pathogen vacuole membrane. mBio 12: e02180‐21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonn, F. , Bartel, J. , Buttner, K. , Hecker, M. , Otto, A. , and Becher, D. (2014) Picking vanished proteins from the void: how to collect and ship/share extremely dilute proteins in a reproducible and highly efficient manner. Anal Chem 86: 7421–7427. [DOI] [PubMed] [Google Scholar]
- Carlson, H.K. , Vance, R.E. , and Marletta, M.A. (2010) H‐NOX regulation of c‐di‐GMP metabolism and biofilm formation in Legionella pneumophila . Mol Microbiol 77: 930–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, B. , Kura, F. , Amemura‐Maekawa, J. , Koizumi, N. , and Watanabe, H. (2005) Identification of a novel adhesion molecule involved in the virulence of Legionella pneumophila . Infect Immun 73: 4272–4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo, S.L. , Bermudez, L.E. , El‐Etr, S.H. , Duhamel, G.E. , and Cirillo, J.D. (2001) Legionella pneumophila entry gene rtxA is involved in virulence. Infect Immun 69: 508–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey, M.E. , and O'Toole, G.A. (2000) Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev 64: 847–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Buck, E. , Maes, L. , Meyen, E. , Van Mellaert, L. , Geukens, N. , Anne, J. , and Lammertyn, E. (2005) Legionella pneumophila Philadelphia‐1 tatB and tatC affect intracellular replication and biofilm formation. Biochem Biophys Res Commun 331: 1413–1420. [DOI] [PubMed] [Google Scholar]
- de Felipe, K.S. , Glover, R.T. , Charpentier, X. , Anderson, O.R. , Reyes, M. , Pericone, C.D. , and Shuman, H.A. (2008) Legionella eukaryotic‐like type IV substrates interfere with organelle trafficking. PLoS Pathog 4: e1000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Declerck, P. (2010) Biofilms: the environmental playground of Legionella pneumophila . Environ Microbiol 12: 557–566. [DOI] [PubMed] [Google Scholar]
- Declerck, P. , Behets, J. , Margineanu, A. , van Hoef, V. , De Keersmaecker, B. , and Ollevier, F. (2009) Replication of Legionella pneumophila in biofilms of water distribution pipes. Microbiol Res 164: 593–603. [DOI] [PubMed] [Google Scholar]
- Declerck, P. , Behets, J. , van Hoef, V. , and Ollevier, F. (2007) Replication of Legionella pneumophila in floating biofilms. Curr Microbiol 55: 435–440. [DOI] [PubMed] [Google Scholar]
- Delgado, M.G. , Rivera, C.A. , and Lennon‐Dumenil, A.M. (2022) Macropinocytosis and cell migration: don't drink and drive. Subcell Biochem 98: 85–102. [DOI] [PubMed] [Google Scholar]
- Dietrich, C. , Heuner, K. , Brand, B.C. , Hacker, J. , and Steinert, M. (2001) Flagellum of Legionella pneumophila positively affects the early phase of infection of eukaryotic host cells. Infect Immun 69: 2116–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyscher, D. , Fieseler, L. , Dons, L. , Loessner, M.J. , and Schuppler, M. (2013) Acanthamoeba feature a unique backpacking strategy to trap and feed on Listeria monocytogenes and other motile bacteria. Environ Microbiol 15: 433–446. [DOI] [PubMed] [Google Scholar]
- Duncan, C. , Prashar, A. , So, J. , Tang, P. , Low, D.E. , Terebiznik, M. , and Guyard, C. (2011) Lcl of Legionella pneumophila is an immunogenic GAG binding adhesin that promotes interactions with lung epithelial cells and plays a crucial role in biofilm formation. Infect Immun 79: 2168–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feeley, J.C. , Gibson, R.J. , Gorman, G.W. , Langford, N.C. , Rasheed, J.K. , Mackel, D.C. , and Baine, W.B. (1979) Charcoal‐yeast extract agar: primary isolation medium for Legionella pneumophila . J Clin Microbiol 10: 437–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields, B.S. (1996) The molecular ecology of Legionellae . Trends Microbiol 4: 286–290. [DOI] [PubMed] [Google Scholar]
- Fields, B.S. , Benson, R.F. , and Besser, R.E. (2002) Legionella and Legionnaires' disease: 25 years of investigation. Clin Microbiol Rev 15: 506–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finsel, I. , and Hilbi, H. (2015) Formation of a pathogen vacuole according to Legionella pneumophila: how to kill one bird with many stones. Cell Microbiol 17: 935–950. [DOI] [PubMed] [Google Scholar]
- Ghosal, D. , Jeong, K.C. , Chang, Y.W. , Gyore, J. , Teng, L. , Gardner, A. , et al. (2019) Molecular architecture, polar targeting and biogenesis of the Legionella Dot/Icm T4SS. Nat Microbiol 4: 1173–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haiko, J. , and Westerlund‐Wikstrom, B. (2013) The role of the bacterial flagellum in adhesion and virulence. Biology 2: 1242–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilbi, H. , Hoffmann, C. , and Harrison, C.F. (2011) Legionella spp. outdoors: colonization, communication and persistence. Environ Microbiol Rep 3: 286–296. [DOI] [PubMed] [Google Scholar]
- Hilbi, H. , Segal, G. , and Shuman, H.A. (2001) Icm/Dot‐dependent upregulation of phagocytosis by Legionella pneumophila . Mol Microbiol 42: 603–617. [DOI] [PubMed] [Google Scholar]
- Hindré, T. , Brüggemann, H. , Buchrieser, C. , and Héchard, Y. (2008) Transcriptional profiling of Legionella pneumophila biofilm cells and the influence of iron on biofilm formation. Microbiology 154: 30–41. [DOI] [PubMed] [Google Scholar]
- Hochstrasser, R. , and Hilbi, H. (2017) Intra‐species and inter‐kingdom signaling of Legionella pneumophila . Front Microbiol 8: 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser, R. , and Hilbi, H. (2019) Migration of Acanthamoeba castellanii through Legionella biofilms. Methods Mol Biol 1921: 79–89. [DOI] [PubMed] [Google Scholar]
- Hochstrasser, R. , and Hilbi, H. (2020) Legionella quorum sensing meets cyclic‐di‐GMP signaling. Curr Opin Microbiol 55: 9–16. [DOI] [PubMed] [Google Scholar]
- Hochstrasser, R. , and Hilbi, H. (2022) The Legionella Lqs‐LvbR regulatory network controls temperature‐dependent growth onset and bacterial cell density. Appl Environ Microbiol 88: e0237021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser, R. , Hutter, C.A.J. , Arnold, F.M. , Barlocher, K. , Seeger, M.A. , and Hilbi, H. (2020) The structure of the Legionella response regulator LqsR reveals amino acids critical for phosphorylation and dimerization. Mol Microbiol 113: 1070–1084. [DOI] [PubMed] [Google Scholar]
- Hochstrasser, R. , Kessler, A. , Sahr, T. , Simon, S. , Schell, U. , Gomez‐Valero, L. , et al. (2019) The pleiotropic Legionella transcription factor LvbR links the Lqs and c‐di‐GMP regulatory networks to control biofilm architecture and virulence. Environ Microbiol 21: 1035–1053. [DOI] [PubMed] [Google Scholar]
- Hoffmann, C. , Harrison, C.F. , and Hilbi, H. (2014) The natural alternative: protozoa as cellular models for Legionella infection. Cell Microbiol 16: 15–26. [DOI] [PubMed] [Google Scholar]
- Horwitz, M.A. (1983) Formation of a novel phagosome by the Legionnaires' disease bacterium (Legionella pneumophila) in human monocytes. J Exp Med 158: 1319–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubber, A. , and Roy, C.R. (2010) Modulation of host cell function by Legionella pneumophila type IV effectors. Annu Rev Cell Dev Biol 26: 261–283. [DOI] [PubMed] [Google Scholar]
- Huws, S.A. , McBain, A.J. , and Gilbert, P. (2005) Protozoan grazing and its impact upon population dynamics in biofilm communities. J Appl Microbiol 98: 238–244. [DOI] [PubMed] [Google Scholar]
- Isberg, R.R. , O'Connor, T.J. , and Heidtman, M. (2009) The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat Rev Microbiol 7: 13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler, A. , Schell, U. , Sahr, T. , Tiaden, A. , Harrison, C. , Buchrieser, C. , and Hilbi, H. (2013) The Legionella pneumophila orphan sensor kinase LqsT regulates competence and pathogen‐host interactions as a component of the LAI‐1 circuit. Environ Microbiol 15: 646–662. [DOI] [PubMed] [Google Scholar]
- Kubori, T. , and Nagai, H. (2016) The type IVB secretion system: an enigmatic chimera. Curr Opin Microbiol 29: 22–29. [DOI] [PubMed] [Google Scholar]
- Kuiper, M.W. , Wullings, B.A. , Akkermans, A.D. , Beumer, R.R. , and van der Kooij, D. (2004) Intracellular proliferation of Legionella pneumophila in Hartmannella vermiformis in aquatic biofilms grown on plasticized polyvinyl chloride. Appl Environ Microbiol 70: 6826–6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau, H.Y. , and Ashbolt, N.J. (2009) The role of biofilms and protozoa in Legionella pathogenesis: implications for drinking water. J Appl Microbiol 107: 368–378. [DOI] [PubMed] [Google Scholar]
- Lucas, C.E. , Brown, E. , and Fields, B.S. (2006) Type IV pili and type II secretion play a limited role in Legionella pneumophila biofilm colonization and retention. Microbiology 152: 3569–3573. [DOI] [PubMed] [Google Scholar]
- Maass, S. , Sievers, S. , Zuhlke, D. , Kuzinski, J. , Sappa, p.K. , Muntel, J. , et al. (2011) Efficient, global‐scale quantification of absolute protein amounts by integration of targeted mass spectrometry and two‐dimensional gel‐based proteomics. Anal Chem 83: 2677–2684. [DOI] [PubMed] [Google Scholar]
- Mallegol, J. , Duncan, C. , Prashar, A. , So, J. , Low, D.E. , Terebeznik, M. , and Guyard, C. (2012) Essential roles and regulation of the Legionella pneumophila collagen‐like adhesin during biofilm formation. PLoS One 7: e46462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mampel, J. , Spirig, T. , Weber, S.S. , Haagensen, J.A.J. , Molin, S. , and Hilbi, H. (2006) Planktonic replication is essential for biofilm formation by Legionella pneumophila in a complex medium under static and dynamic flow conditions. Appl Environ Microbiol 72: 2885–2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manske, C. , Schell, U. , and Hilbi, H. (2016) Metabolism of myo‐inositol by Legionella pneumophila promotes infection of amoebae and macrophages. Appl Environ Microbiol 82: 5000–5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matz, C. , and Kjelleberg, S. (2005) Off the hook – how bacteria survive protozoan grazing. Trends Microbiol 13: 302–307. [DOI] [PubMed] [Google Scholar]
- Moffat, J.F. , and Tompkins, L.S. (1992) A quantitative model of intracellular growth of Legionella pneumophila in Acanthamoeba castellanii . Infect Immun 60: 296–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murga, R. , Forster, T.S. , Brown, E. , Pruckler, J.M. , Fields, B.S. , and Donlan, R.M. (2001) Role of biofilms in the survival of Legionella pneumophila in a model potable‐water system. Microbiology 147: 3121–3126. [DOI] [PubMed] [Google Scholar]
- Nagai, H. , Kagan, J.C. , Zhu, X. , Kahn, R.A. , and Roy, C.R. (2002) A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science 295: 679–682. [DOI] [PubMed] [Google Scholar]
- Neunuebel, M.R. , Chen, Y. , Gaspar, A.H. , Backlund, p.S., Jr. , Yergey, A. , and Machner, M.P. (2011) De‐AMPylation of the small GTPase Rab1 by the pathogen Legionella pneumophila . Science 333: 453–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton, H.J. , Ang, D.K. , van Driel, I.R. , and Hartland, E.L. (2010) Molecular pathogenesis of infections caused by Legionella pneumophila . Clin Microbiol Rev 23: 274–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pécastaings, S. , Allombert, J. , Lajoie, B. , Doublet, P. , Roques, C. , and Vianney, A. (2016) New insights into Legionella pneumophila biofilm regulation by c‐di‐GMP signaling. Biofouling 32: 935–948. [DOI] [PubMed] [Google Scholar]
- Pécastaings, S. , Berge, M. , Dubourg, K.M. , and Roques, C. (2010) Sessile Legionella pneumophila is able to grow on surfaces and generate structured monospecies biofilms. Biofouling 26: 809–819. [DOI] [PubMed] [Google Scholar]
- Perez‐Riverol, Y. , Csordas, A. , Bai, J. , Bernal‐Llinares, M. , Hewapathirana, S. , Kundu, D.J. , et al. (2019) The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 47: D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Personnic, N. , Bärlocher, K. , Finsel, I. , and Hilbi, H. (2016) Subversion of retrograde trafficking by translocated pathogen effectors. Trends Microbiol 24: 450–462. [DOI] [PubMed] [Google Scholar]
- Personnic, N. , Striednig, B. , and Hilbi, H. (2018) Legionella quorum sensing and its role in pathogen‐host interactions. Curr Opin Microbiol 41: 29–35. [DOI] [PubMed] [Google Scholar]
- Personnic, N. , Striednig, B. , and Hilbi, H. (2021) Quorum sensing controls persistence, resuscitation, and virulence of Legionella subpopulations in biofilms. ISME J 15: 196–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Personnic, N. , Striednig, B. , Lezan, E. , Manske, C. , Welin, A. , Schmidt, A. , and Hilbi, H. (2019) Quorum sensing modulates the formation of virulent Legionella persisters within infected cells. Nat Commun 10: 5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova, O.E. , and Sauer, K. (2012) Sticky situations: key components that control bacterial surface attachment. J Bacteriol 194: 2413–2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, J. , and Luo, Z.Q. (2017) Legionella and Coxiella effectors: strength in diversity and activity. Nat Rev Microbiol 15: 591–605. [DOI] [PubMed] [Google Scholar]
- Qiu, J. , Sheedlo, M.J. , Yu, K. , Tan, Y. , Nakayasu, E.S. , Das, C. , et al. (2016) Ubiquitination independent of E1 and E2 enzymes by bacterial effectors. Nature 533: 120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronn, R. , McCaig, A.E. , Griffiths, B.S. , and Prosser, J.I. (2002) Impact of protozoan grazing on bacterial community structure in soil microcosms. Appl Environ Microbiol 68: 6094–6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothmeier, E. , Pfaffinger, G. , Hoffmann, C. , Harrison, C.F. , Grabmayr, H. , Repnik, U. , et al. (2013) Activation of Ran GTPase by a Legionella effector promotes microtubule polymerization, pathogen vacuole motility and infection. PLoS Pathog 9: e1003598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadosky, A.B. , Wiater, L.A. , and Shuman, H.A. (1993) Identification of Legionella pneumophila genes required for growth within and killing of human macrophages. Infect Immun 61: 5361–5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell, U. , Kessler, A. , and Hilbi, H. (2014) Phosphorylation signalling through the Legionella quorum sensing histidine kinases LqsS and LqsT converges on the response regulator LqsR. Mol Microbiol 92: 1039–1055. [DOI] [PubMed] [Google Scholar]
- Schell, U. , Simon, S. , Sahr, T. , Hager, D. , Albers, M.F. , Kessler, A. , et al. (2016) The α‐hydroxyketone LAI‐1 regulates motility, Lqs‐dependent phosphorylation signalling and gene expression of Legionella pneumophila . Mol Microbiol 99: 778–793. [DOI] [PubMed] [Google Scholar]
- Segal, G. , Russo, J.J. , and Shuman, H.A. (1999) Relationships between a new type IV secretion system and the icm/dot virulence system of Legionella pneumophila . Mol Microbiol 34: 799–809. [DOI] [PubMed] [Google Scholar]
- Segal, G. , and Shuman, H.A. (1998) Intracellular multiplication and human macrophage killing by Legionella pneumophila are inhibited by conjugal components of IncQ plasmid RSF1010. Mol Microbiol 30: 197–208. [DOI] [PubMed] [Google Scholar]
- Segal, G. , and Shuman, H.A. (1999) Legionella pneumophila utilizes the same genes to multiply within Acanthamoeba castellanii and human macrophages. Infect Immun 67: 2117–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon, S. , Schell, U. , Heuer, N. , Hager, D. , Albers, M.F. , Matthias, J. , et al. (2015) Inter‐kingdom signaling by the Legionella quorum sensing molecule LAI‐1 modulates cell migration through an IQGAP1‐Cdc42‐ARHGEF9‐dependent pathway. PLoS Pathog 11: e1005307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon, S. , Wagner, M.A. , Rothmeier, E. , Müller‐Taubenberger, A. , and Hilbi, H. (2014) Icm/Dot‐dependent inhibition of phagocyte migration by Legionella is antagonized by a translocated Ran GTPase activator. Cell Microbiol 16: 977–992. [DOI] [PubMed] [Google Scholar]
- Spirig, T. , Tiaden, A. , Kiefer, P. , Buchrieser, C. , Vorholt, J.A. , and Hilbi, H. (2008) The Legionella autoinducer synthase LqsA produces an α‐hydroxyketone signaling molecule. J Biol Chem 283: 18113–18123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreelatha, A. , Nolan, C. , Park, B.C. , Pawlowski, K. , Tomchick, D.R. , and Tagliabracci, V.S. (2020) A Legionella effector kinase is activated by host inositol hexakisphosphate. J Biol Chem 295: 6214–6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner, B. , Swart, A.L. , Welin, A. , Weber, S. , Personnic, N. , Kaech, A. , et al. (2017) ER remodeling by the large GTPase atlastin promotes vacuolar growth of Legionella pneumophila . EMBO Rep 18: 1817–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner, B. , Weber, S. , and Hilbi, H. (2018) Formation of the Legionella‐containing vacuole: phosphoinositide conversion, GTPase modulation and ER dynamics. Int J Med Microbiol 308: 49–57. [DOI] [PubMed] [Google Scholar]
- Stewart, C.R. , Muthye, V. , and Cianciotto, N.P. (2012) Legionella pneumophila persists within biofilms formed by Klebsiella pneumoniae, Flavobacterium sp., and Pseudomonas fluorescens under dynamic flow conditions. PLoS One 7: e50560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Striednig, B. , Lanner, U. , Niggli, S. , Katic, A. , Vormittag, S. , Brülisauer, S. , et al. (2021) Quorum sensing governs a transmissive Legionella subpopulation at the pathogen vacuole periphery. EMBO Rep 22: e52972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swart, A.L. , Gomez‐Valero, L. , Buchrieser, C. , and Hilbi, H. (2020a) Evolution and function of bacterial RCC1 repeat effectors. Cell Microbiol 22: e13246. [DOI] [PubMed] [Google Scholar]
- Swart, A.L. , Harrison, C.F. , Eichinger, L. , Steinert, M. , and Hilbi, H. (2018) Acanthamoeba and Dictyostelium as cellular models for Legionella infection. Front Cell Infect Microbiol 8: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swart, A.L. , Steiner, B. , Gomez‐Valero, L. , Schutz, S. , Hannemann, M. , Janning, P. , et al. (2020b) Divergent evolution of Legionella RCC1 repeat effectors defines the range of Ran GTPase cycle targets. mBio 11: e00405‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, Y. , and Luo, Z.Q. (2011) Legionella pneumophila SidD is a deAMPylase that modifies Rab1. Nature 475: 506–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiaden, A. , and Hilbi, H. (2012) α‐Hydroxyketone synthesis and sensing by Legionella and Vibrio . Sensors 12: 2899–2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiaden, A. , Spirig, T. , Carranza, P. , Brüggemann, H. , Riedel, K. , Eberl, L. , et al. (2008) Synergistic contribution of the Legionella pneumophila lqs genes to pathogen‐host interactions. J Bacteriol 190: 7532–7547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiaden, A. , Spirig, T. , and Hilbi, H. (2010a) Bacterial gene regulation by α‐hydroxyketone signaling. Trends Microbiol 18: 288–297. [DOI] [PubMed] [Google Scholar]
- Tiaden, A. , Spirig, T. , Sahr, T. , Wälti, M.A. , Boucke, K. , Buchrieser, C. , and Hilbi, H. (2010b) The autoinducer synthase LqsA and putative sensor kinase LqsS regulate phagocyte interactions, extracellular filaments and a genomic Island of Legionella pneumophila . Environ Microbiol 12: 1243–1259. [DOI] [PubMed] [Google Scholar]
- Tiaden, A. , Spirig, T. , Weber, S.S. , Brüggemann, H. , Bosshard, R. , Buchrieser, C. , and Hilbi, H. (2007) The Legionella pneumophila response regulator LqsR promotes host cell interactions as an element of the virulence regulatory network controlled by RpoS and LetA. Cell Microbiol 9: 2903–2920. [DOI] [PubMed] [Google Scholar]
- Vandersmissen, L. , De Buck, E. , Saels, V. , Coil, D.A. , and Anne, J. (2010) A Legionella pneumophila collagen‐like protein encoded by a gene with a variable number of tandem repeats is involved in the adherence and invasion of host cells. FEMS Microbiol Lett 306: 168–176. [DOI] [PubMed] [Google Scholar]
- Veltman, D.M. (2015) Drink or drive: competition between macropinocytosis and cell migration. Biochem Soc Trans 43: 129–132. [DOI] [PubMed] [Google Scholar]
- Vizcaino, J.A. , Csordas, A. , del‐Toro, N. , Dianes, J.A. , Griss, J. , Lavidas, I. , et al. (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res 44: D447–D456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, S.S. , Joller, N. , Kuntzel, A.B. , Spörri, R. , Tchang, V.S. , Scandella, E. , et al. (2012) Identification of protective B cell antigens of Legionella pneumophila . J Immunol 189: 841–849. [DOI] [PubMed] [Google Scholar]
- Weber, S.S. , Ragaz, C. , Reus, K. , Nyfeler, Y. , and Hilbi, H. (2006) Legionella pneumophila exploits PI(4)P to anchor secreted effector proteins to the replicative vacuole. PLoS Pathog 2: e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Heat map of proteins depleted or enriched in ∆lvbR and ∆lqsR biofilms compared to JR32.
Comparative proteomics was performed with biofilms formed by L. pneumophila JR32, ΔlvbR or ΔlqsR. In the heat map all proteins are shown that are depleted or enriched in mutant biofilms (for details see Materials and Methods). Biofilm bacteria were harvested by centrifugation and supernatants (designated ‘s’, extracellular) and cell pellets (designated ‘p’, intracellular) were analysed separately. The proteins are listed by ascending Lpg numbers. The colour code indicates the level of regulation: the darker the colour the stronger the corresponding gene is regulated (red: depleted proteins, max. 2−5; blue: accumulated proteins, max. 27.4). Boxes highlight regions of interest, which are further discussed. ‘NaN’: not a number.
Fig. S2. Biofilm formation and quantification of L. pneumophila strains.
(A) Biofilms were initiated with exponential phase L. pneumophila ∆lqsS, ∆lqsT or ∆lqsS‐∆lqsT mutant strains harbouring pNT28 (constitutive production of GFP) in AYE medium within ibiTreat microscopy dishes. Biofilms were grown at 25°C without mechanical disturbance and confocal microscopy images of biofilm architecture were acquired at 4 μm above dish bottom after 1, 2, 3 and 6 days of growth. The images shown are representative of at least two independent experiments. Scale bars, 30 μm. (B, C) Quantification of L. pneumophila biofilms grown for 6 days. (B) Pseudocolour graph (GFP intensity vs. FSC) depicting the gate for GFP‐producing L. pneumophila (identified by fluorescence) and the beads population (identified by SSC vs. FSC). (C) Means and standard deviation (bacteria/ml) of biological triplicates from 100′000 events of each biofilm suspension (differences between wild‐type and mutant L. pneumophila are statistically not significant).
Fig. S3. Surface adherence of biofilms formed by L. pneumophila JR32 and mutant strains.
Biofilms were initiated with exponential phase L. pneumophila JR32, ∆lvbR, ∆lqsR, ∆lqsA, ∆lqsS, ∆lqsT, ∆lqsS‐∆lqsT, ∆flaA or ∆icmT mutant strains harbouring pNT28 (constitutive production of GFP) in AYE medium within ibiTreat microscopy dishes. Biofilms were grown at 25°C without mechanical disturbance and confocal microcopy images of biofilm attachments were acquired at the dish bottom (0 μm) after 1, 2, 3 and 6 days of growth. The images shown are representative of at least two independent experiments. Scale bars, 10 μm.
Fig. S4. L. pneumophila cluster formation on A. castellanii in JR32 or mutant strain biofilms.
Biofilms of (A) L. pneumophila JR32, ∆lvbR, ∆lqsR, ∆lqsA, ∆flaA or ∆icmT mutant strains harbouring pNT28 (constitutive production of GFP), or (B, C) strain JR32 harbouring pNT28 were grown for 6 days in AYE medium at 25°C, and A. castellanii was added to preformed biofilms. Bacterial adherence and cluster formation were monitored by confocal microscopy (A, B) ca. 4 μm above or (C) at the dish bottom ("clusters": arrow heads, "trails": arrows). (D, E) Biofilms of L. pneumophila JR32 or ∆flaA mutant strains constitutively producing GFP (pNT28) or GFP and FlaA under control of its promoter (∆flaA/+flaA, pSB001) were grown for 6 days in AYE medium at 25°C, and A. castellanii was added to preformed biofilms. Confocal microscopy images of (D) overview and (E) zoom‐in of L. pneumophila adherence and cluster formation on amoebae. The images shown are representative of three independent experiments. Scale bars, 30 μm.
Fig. S5. Quantification of amoeba‐associated L. pneumophila clusters by flow cytometry.
Quantification by flow cytometry (forward scatter area, FSC‐A, vs. fluorescence intensity) using FlowJo software of (A) L. pneumophila strain JR32 or A. castellanii, (B) amoebae‐associated or non‐associated, GFP‐positive L. pneumophila JR32, ∆lvbR, ∆lqsR, ∆lqsA, ∆flaA or ∆icmT harbouring pNT28 (protocol #1, see Material and Methods), or (C) non‐associated, GFP‐positive L. pneumophila JR32 harbouring promoter expression reporters (P flaA , P 6SRNA , P sidC , P ralF , or P csrA ) (protocol #2, see Material and Methods).
Fig. S6. Competition of mCherry‐producing L. pneumophila JR32 and GFP‐producing mutants for cluster formation on amoebae.
Biofilm formation was initiated with a 1:1 ratio of exponential phase mCherry‐producing L. pneumophila JR32 (pNP102) and GFP‐producing JR32 or mutant strains (pNT28). Biofilms were grown in AYE medium within ibiTreat microscopy dishes at 25°C for 6 days. A. castellanii were added to preformed biofilms, and amoebae with adherent bacterial clusters were monitored by confocal microcopy above dish bottom. The images shown are representative of at least two independent experiments. Scale bars, 30 μm.
Fig. S7. Gene expression kinetics of L. pneumophila JR32 in AYE medium using single promoter reporters.
L. pneumophila JR32 harbouring (A) P flaA ‐gfp (pCM009), (B) P 6SRNA ‐gfp (pRH049), (C) P sidC ‐gfp (pRH035), (D) P ralF ‐gfp (pRH032) or (E) P csrA ‐gfp (pRH031) reporter constructs were grown at 37°C in AYE medium in microplates while orbitally shaking. GFP fluorescence (relative fluorescence units, RFU) at a gain of 50 and optical density at 600 nm (OD600) were monitored over time using a microplate reader. The kinetics of the GFP fluorescence or OD600 values are shown and depicted by the left or right y‐axis respectively. The data are means and standard deviations of technical triplicates and representative of three independent measurements.
Fig. S8. Gene expression kinetics of L. pneumophila JR32 in AYE medium and in amoeba‐adherent bacteria in biofilms using dual promoter reporters.
L. pneumophila JR32 harbouring (A) P flaA ‐gfp (pSN7) or (B) P 6SRNA ‐gfp (pSN25) reporter constructs were grown at 37°C in AYE medium within microplates while orbitally shaking. GFP fluorescence (relative fluorescence units, RFU) at a gain of 50 and optical density at 600 nm (OD600) were monitored over time using a microplate reader. The kinetics of the GFP fluorescence (left y‐axis) or OD600 (right y‐axis) values are shown. The data are means and standard deviations of technical triplicates and representative of three independent measurements. (C, D) Biofilms of L. pneumophila JR32, ∆lvbR or ∆lqsR harbouring dual reporter plasmids expressing P tac ‐mcherry (constitutive) and P flaA ‐gfp (pSN7) or P 6SRNA ‐gfp (pSN25) were grown in AYE medium for 6 days. A. castellanii was added to preformed biofilms, and confocal microcopy images of amoebae with adherent bacterial clusters were acquired close to the bottom of microscopy dishes. The images shown are representative of 2–3 independent experiments. Scale bars, 30 μm.
Table S1. Oligonucleotides used in this study.
Movie S1. Motility of L. pneumophila JR32.
Movie S2. Motility of L. pneumophila ∆lvbR.
Movie S3. Motility of L. pneumophila ∆lqsR.
Movie S4. A. castellanii migrating through JR32 biofilm.
Movie S5. A. castellanii migrating through ∆lvbR biofilm.
Movie S6. A. castellanii migrating through ∆lqsR biofilm.
Movie S7. A. castellanii migrating through ∆lqsA biofilm.
Movie S8. A. castellanii migrating through ∆flaA biofilm.
Movie S9. A. castellanii migrating through ∆icmT biofilm.
Data Availability Statement
The MS proteomics data discussed in this publication have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (Vizcaino et al., 2016; Perez‐Riverol et al., 2019) with the dataset identifier PXD030337 (Reviewer account details: Username, reviewer_pxd030337@ebi.ac.uk; Password, a2Q4ygNx).


