Figure 1.
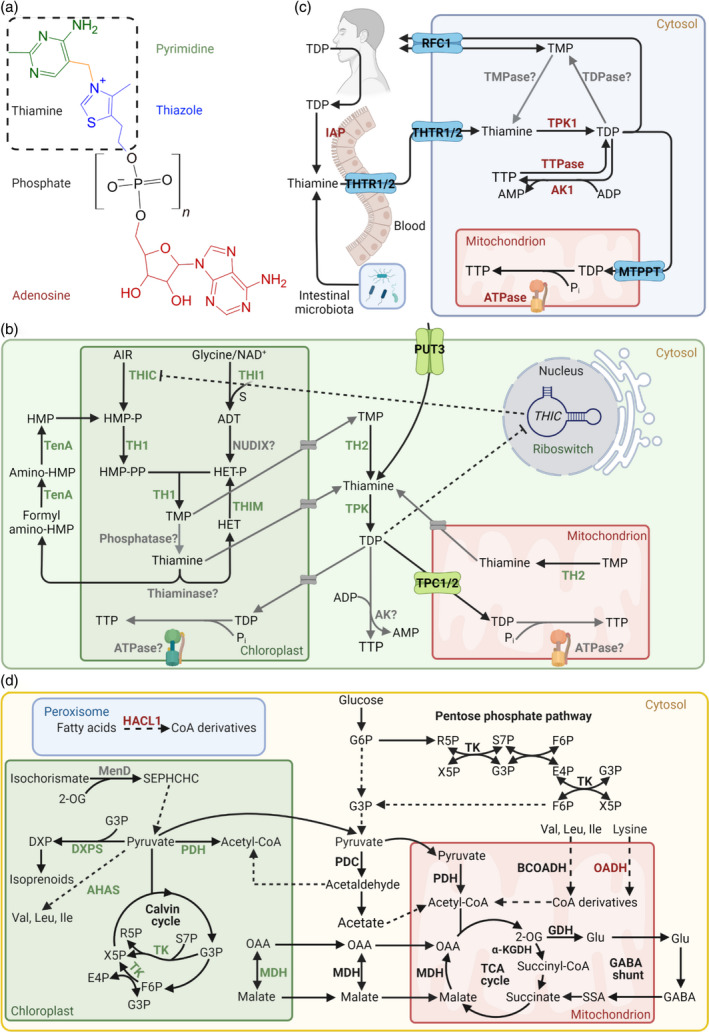
Vitamin B1 metabolism in plants and humans. (a) Generalized chemical structure of vitamin B1. The basic unit thiamine (hashed box) consists of a pyrimidine (green) and thiazole (blue) heterocycle, bridged by a methylene group (yellow). Thiamine derivatives vary in phosphorylation states (black, n = 1–3), as thiamine monophosphate (TMP), thiamine diphosphate (TDP), thiamine triphosphate (TTP), and in adenosylation states (red), as adenosine TDP (ATDP) and adenosine TTP (ATTP). (b) Vitamin B1 metabolism in plants. Biosynthesis de novo takes place in the chloroplast, where each heterocycle is separately synthesized. 5‐Aminoimidazole ribonucleotide (AIR) is used by THIC to generate 4‐amino‐5‐hydroxymethyl‐2‐methylpyrimidine phosphate (HMP‐P) and is further phosphorylated by TH1 to generate 4‐amino‐5‐hydroxymethyl‐2‐methylpyrimidine pyrophosphate (HMP‐PP). Glycine, nicotinamide adenine dinucleotide (NAD+) and a sulfur from a cysteine residue in the THI1 backbone is used to generate the adenylated thiazole intermediate (ADT) and is hydrolyzed into 4‐methyl‐5‐(2‐hydroxyethyl)thiazole phosphate HET‐P by a nucleoside diphosphate hydrolase (NUDIX, gray), but remains to be characterized. The two heterocycles are condensed by TH1 into TMP, which is dephosphorylated to thiamine by a phosphatase, although it is not clear if this can take place in the chloroplast. In any case, TH2 in the cytosol and mitochondrion can dephosphorylate TMP to thiamine, which is then pyrophosphorylated to the coenzyme form TDP by thiamine pyrophosphokinase (TPK) in the cytosol. Cytosolic TDP must be transported back into the chloroplast and mitochondrion for use in enzyme reactions (d). Although most vitamin B1 transporters remain to be identified (putative transporters depicted in gray), polyamine uptake transporter 3 (PUT3, green) facilitates thiamine transport through the plasma membrane and the thiamine phosphate carriers (TPC1/2, green) facilitate TDP transport into the mitochondrion. The biosynthesis of TTP in plants remains to be fully elucidated but may be derived from TDP by adenylate kinase (AK) in the cytosol or through ATP synthase (ATPase) in the chloroplast and mitochondrion. Regulation of TDP biosynthesis occurs through TDP binding to the THIC riboswitch in the nucleus, which in turn downregulates THIC expression (dashed lines). Thiamine can also be catabolized to derivatives of its respective heterocycles by thiaminase (and needs further investigation in plants), which can be recycled through TenA and THIM. In the pyrimidine branch, TenA can hydrolyze both formylamino‐HMP and amino‐HMP into HMP. In the thiazole branch, THIM has kinase activity and phosphorylates HET into HET‐P. (c) Metabolism of vitamin B1 in humans. Humans depend on dietary uptake and absorption of vitamin B1, the most abundant form of which is TDP and is converted into thiamine by the intestinal alkaline phosphatase (IAP) for uptake by thiamine transporter 1/2 (THTR1/2). Intestinal microbiota can also produce thiamine for human usage. THTR1/2 facilitate cellular thiamine uptake, which is pyrophosphorylated in the cytosol to TDP by TPK1. The reduced folate carrier 1 (RFC1) can facilitate the transport of TDP and TMP across the plasma membrane and the mitochondrial thiamine pyrophosphate transporter (MTPPT) can transport TDP into the mitochondrion. The dephosphorylation of TDP by a thiamine pyrophosphatase (TDPase) and TMP by a thiamine monophosphatase (TMPase) occurs inside the cell, but the precise genes are not known. TTP biosynthesis is well characterized in humans and is postulated to occur through adenylate kinase 1 (AK1) in the cytosol using TDP and adenosine diphosphate (ADP) as substrates, and through the mitochondrial ATPase using TDP and inorganic phosphate (Pi) as substrates. Cytosolic thiamine triphosphatase (TTPase) is responsible for TTP hydrolysis to TDP. (d) An overview of enzymatic reactions that either use TDP as a coenzyme or are allosterically regulated by B1 vitamers. Enzymes specific for plants and humans are highlighted in green and red, respectively, and those common to both are presented in black. All enzymes shown here (bold) are TDP‐dependent except GDH (allosterically regulated by TTP and ATTP) and MDH (allosterically regulated by TDP and thiamine). MenD (gray) has a TDP‐dependent feature in microorganisms and its plant homolog has a TDP‐binding domain but has not undergone investigation. Abbreviations: Acetyl‐CoA, acetyl coenzyme A; AHAS, acetohydroxyacid synthase; BCOADH, branched chain oxy‐acid dehydrogenase; DXP, 1‐deoxy‐d‐xylulose‐5‐phosphate; DXPS, DXP synthase; E4P, erythrose‐4‐phosphate; F6P, fructose‐6‐phosphate; GABA, γ‐amino‐butyric acid; Glu, glutamate; G3P, glyceraldehyde 3‐phosphate; G6P, glucose 6‐phosphate; HACL1, 2‐hydroxyacyl‐CoA lyase 1; α‐KGDH, α‐ketoglutarate dehydrogenase; MDH, malate dehydrogenase; MenD, SEPHCHC synthase; OAA, oxaloacetate; OADH, 2‐oxoadipate dehydrogenase; 2‐OG, α‐ketoglutarate or 2‐oxoglutarate; PDC, pyruvate decarboxylase; PDH, pyruvate dehydrogenase; R5P, ribose 5‐phosphate; SEPHCHC, 5‐enolpyruvoyl‐6‐hydroxy‐2‐succinyl‐cyclohex‐3‐ene‐1‐carboxylate; S7P, sedoheptulose‐7‐phosphate; SSA, succinic semialdehyde; TK, transketolase; X5P, xylulose‐5‐phosphate. [Colour figure can be viewed at wileyonlinelibrary.com]
