Figure 2.
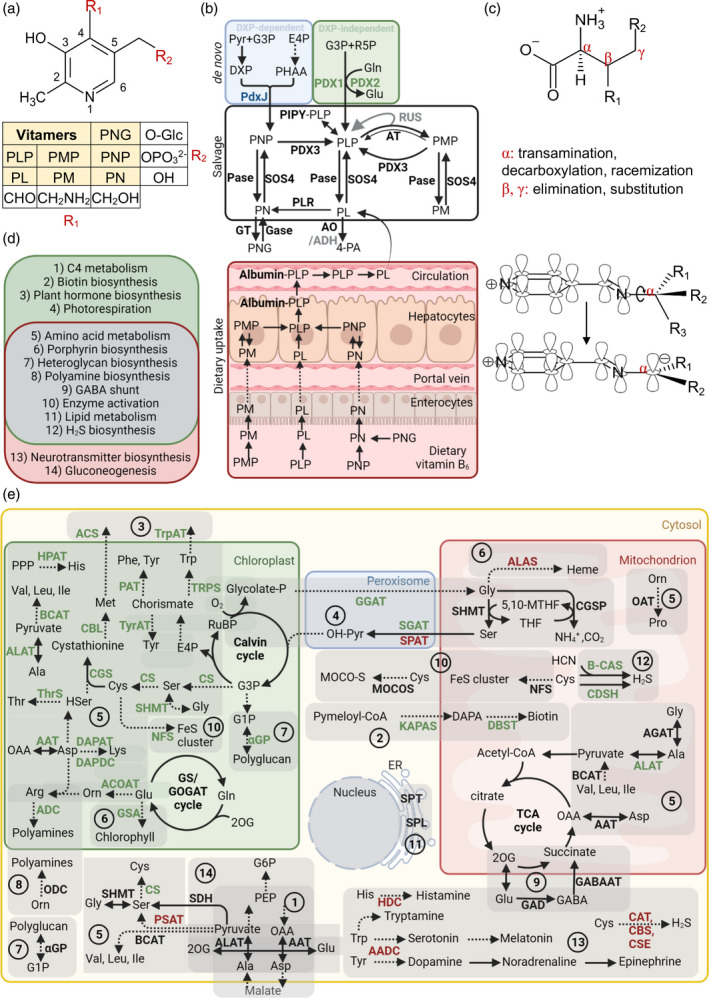
Vitamin B6 metabolism in plants and humans. (a) Generalized chemical structure of vitamin B6. Functional groups at C4 (R1) and C5 (R2) corresponding to the different vitamers are indicated in the box below. Vitamers are pyridoxal (PL), pyridoxamine (PM), pyridoxine (PN) and their phosphorylated derivatives, pyridoxal 5′‐phosphate (PLP), pyridoxamine 5′‐phosphate (PMP), pyridoxine 5′‐phosphate (PNP) and pyridoxine‐5′‐β‐d‐glucoside (PNG). (b) Vitamin B6 can be biosynthesized de novo via two pathways. The DXP‐dependent pathway (blue background) used by certain bacteria generates PNP from deoxyxylulose 5‐phosphate (DXP) and 3‐phosphohydroxy‐1‐aminoacetone (PHAA). DXP is formed from pyruvate and glyceraldehyde 3‐phosphate (G3P), and PHAA is generated through a chain of reactions from erythrose‐4‐phosphate (E4P). In the DXP‐independent pathway (green background), used by plants, PDX1 generates PLP directly from G3P and ribose 5‐phosphate (R5P), with PDX2 providing ammonia from glutamine. The salvage pathway regulates the balance between the different vitamer forms and contributes to vitamin B6 uptake in animals (bottom panel). Phosphatases (Pase) and the pyridoxal kinase (SOS4) are responsible for the interconversion of the phosphorylated and dephosphorylated vitamers, respectively. Pyridoxal reductase (PLR) can reduce the PL pool by converting it to PN. The storage forms of vitamin B6 in plants are glucosylated derivatives of PN, such as PNG, generated by glycosyltransferases (GT), and the remobilization of PNG into PN is catalyzed by glucosidases (Gase). Aminotransferases (AT) are responsible for the generation of PMP from PLP by a mechanism in which PMP dissociates from the enzyme after the first transamination half‐reaction (represented by a thicker arrow). This free PMP can be converted back to PLP by the aminotransferases (represented by a thinner arrow) or by the PMP/PNP oxidase (PDX3). Certain aminotransferases are proposed to regulate free PLP levels by sequestering PLP, which is reversed by interactions with RUS1 and RUS2 (RUS). The PLP homeostasis protein (PIPY) binds PLP covalently and is involved in balancing the different vitamer forms by unknown mechanisms. The degradation of vitamin B6 is catalyzed by aldehyde oxidases (AO) that generate 4‐pyridoxic acid (4‐PA) from PL. NAD‐dependent aldehyde dehydrogenases (ADH) are also implicated in this process. Factors in gray deserve broader investigation. (c) Type of reactions at given positions on an amino acid substrate that are catalyzed by PLP. Dunathan's hypothesis showing a simplified structure of PLP in a Schiff‐base link that enables the deprotonation or decarboxylation of the α‐carbon of the substrate amino acid by providing a delocalization path to the electron of the resulting carbanion is represented below. (d) Physiological roles of PLP as a coenzyme in plants (green) and humans (pink), with overlapping roles listed in the middle (gray). (e) An overview of enzymatic reactions that use PLP as a coenzyme in plants and humans, with their subcellular localization and the physiological processes that they are involved in labeled using the numbering from panel (d) and grouped with a light‐gray shadow. Enzymes specific for plants and humans are highlighted in green and red, respectively, and enzymes common to both are in black. Abbreviations: AADC, aromatic amino acid decarboxylase; AAT, aspartate aminotransferase; ACOAT, acetylornithine aminotransferase; ACS, 1‐aminocyclopropane‐1‐carboxylate (ACC) synthase; ADC, arginine decarboxylase; ALAS, 5‐aminolevulinate synthase; ALAT, alanine aminotransferase; B‐CAS, β‐cyanoalanine synthase; BCAT, branched‐chain amino acid aminotransferase; CAT, cysteine aminotransferase; CBL, cystathionine‐β‐lyase; CBS, cystathionine‐β‐synthase; CDSH, cysteine desulfhydrase; CGS, cystathionine‐γ‐synthase; CS, cysteine synthase; CSE, cystathionine‐γ‐lyase; DAPAT, diaminopimelate aminotransferase; DAPDC, diaminopimelate decarboxylase; DTBS, dethiobiotin synthetase; GABA‐AT, γ‐amino‐butyric acid aminotransferase; GAD, glutamate decarboxylase; GCSP, glycine cleavage system P protein; GGAT, glutamate‐glyoxalate aminotransferase; αGP, α‐glucan phosphorylase; GSA, glutamate‐1‐semialdehyde 2,1‐aminomutase; HDC, histidine decarboxylase; HPAT, histidinolphosphate aminotransferase; KAPAS, 7‐keto‐8‐amino‐pelargonic acid synthase; MOCOS, molybdenum cofactor sulphurase; NFS, cysteine desulphurase; OAT, ornithine aminotransferase; ODC, ornithine decarboxylase; PAT, prephenate aminotransferase; PSAT, phosphoserine aminotransferase; SDH, serine dehydratase; SGAT, serine‐glyoxalate aminotransferase; SHMT, serine hydroxamethyltransferase; SPAT, serine‐pyruvate aminotransferase; SPL, sphingosine‐1‐phosphate lyase; SPT, serine palmitoyltransferase; ThrS, threonine synthase; TrpAT, tryptophan aminotransferase; TRPS, tryptophan synthase; TyrAT, tyrosine aminotransferase. [Colour figure can be viewed at wileyonlinelibrary.com]
