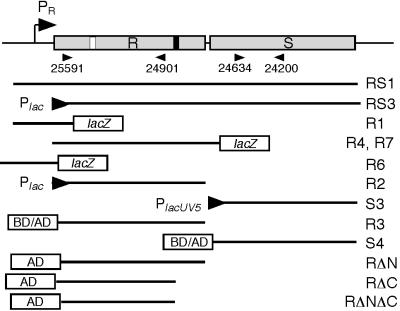
FIG. 1.
Features of the hrpRS region and primers and constructs used in experiments. A map of the hrpRS region is shown at the top. Shaded boxes represent deduced coding sequences for hrpR (R) and hrpS (S) (9, 63). The location of the hrpR promoter (Pr) is shown by the bent arrow. The HrpR box (white bar) and hrpS promoter (black bar) are positioned as proposed by Grimm et al. (15). The locations and orientations of the primers used in the RT-PCR experiments are indicated by the labeled arrowheads immediately below the map. Fragments used in other experiments are indicated by the labeled lines that represent the portion of the mapped region carried by the fragment. Promoter fusions are shown by the arrowheads, with the left side representing the left end of the fragment. Transcriptional fusions to lacZ are indicated by the labeled box. Translational fusions constructed to the LexA′ BD or GAL4′ AD are also shown. Not shown is the S23 fragment that is equivalent to the S4 fragment except that it has tails with different restriction sites; this fragment was used to construct a His-tagged derivative as described in Materials and Methods. The R8 fragment is equivalent to the R3 fragment but lacks the C-terminal stop codon and was used to make a FLAG-tagged derivative. Designations for plasmids carrying the above-described fragments represent the constructor's initials, the fragment designation shown above, and a suffix indicating the host vector. Vector codes are as follows: B, pBluescript SK+; BTM, pBTM116; CTC, pFLAG-CTC; D, pDSK519; DS, pDSK600; GAD, pGAD424; L, pLAFR3; MS, pMLB1034; Q30, pQE30; R, pRG970.