Figure 1.
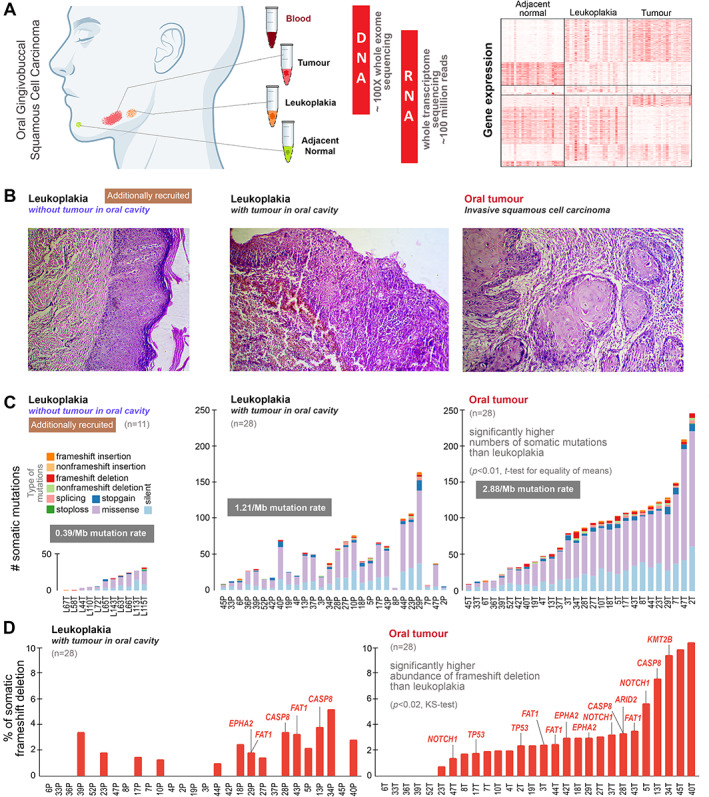
(A) To understand the genomic underpinnings of malignant progression of oral potentially malignant lesion (leukoplakia), oral cancer patients were recruited in whom there was the presence of leukoplakia with cancer. From each patient, tissue samples from tumour, leukoplakia, adjacent normal, and blood were collected. Good‐quality DNA was extracted from tumour, leukoplakia, and blood and sequenced for whole‐exome at about 100X depth; RNA was extracted from tumour, leukoplakia, and adjacent normal tissue and whole transcriptome sequence data were generated (the right‐hand portion represents mRNA expression patterns of differentially expressed genes in leukoplakia and tumour with respect to adjacent normal tissue, described in the supplementary information). (B) Additionally, we recruited 11 individuals diagnosed having oral leukoplakia without the presence of oral tumour. The left‐hand‐side H&E‐stained image showing a leukoplakia lesion in squamous epithelium with features of dysplasia in patients without the presence of tumour in the oral cavity. The middle image represents the histopathology of leukoplakia with the concomitant presence of tumour within the oral cavity, revealing progression from mildly dysplastic epithelium to invasive squamous cell carcinoma with infiltrating dyscohesive neoplastic epithelial cells. And the right‐hand‐side image shows the histopathological features of invasive squamous cell carcinoma with invading neoplastic squamous cells islands in connective tissue stroma. (C) The numbers of somatic mutations in leukoplakia of 11 individuals without tumour in the oral cavity, and in both leukoplakia and tumour in 28 patients with concomitant leukoplakia and cancer are provided. The somatic mutation rate in oral tumours (2.88/Mb) was higher than in leukoplakia (1.21/Mb). The mean number of somatic mutations in tumours (84) was significantly higher than in leukoplakia lesions (44) of the same patient. Additionally, we generated whole‐exome sequence data from the oral leukoplakia tissue of 11 individuals without oral cancer, among whom the mean number of somatic mutations (14) was smaller. (D) The proportion of frameshift deletions in tumours was significantly higher than in leukoplakia tissue. Frameshift deletions in known oral cancer driver genes are provided for each patient.
