FIGURE 2.
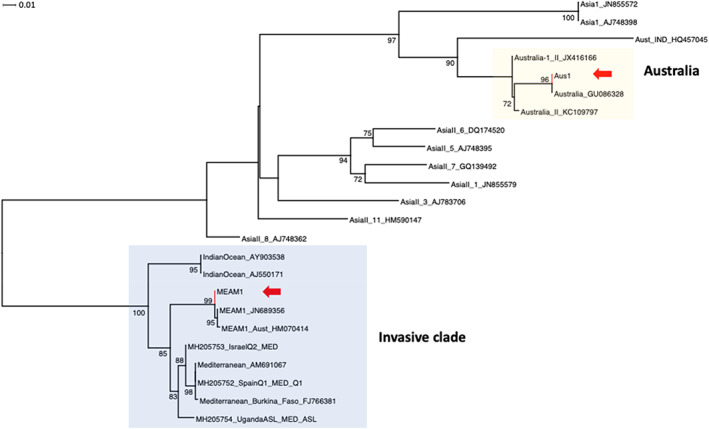
An unrooted maximum‐likelihood (ML) phylogenetic tree based on 483 bp of Bemisia tabaci partial mtCOI gene sequences using IQTree. 48 Arrows show the phylogenetic placements (with high node support values) of the two cryptic species (AUS I, MEAM1) detected in this study against selected partial mtCOI gene sequences 13 of species including the other endemic AUS II species (JX416166, KC109797), an Indonesian species (HQ457045), various Asian species, the Indian Ocean, Mediterranean (MED) species complex and the MEAM1 species. 9 Node confidence estimates are based on 1000 UltraFast bootstrap replications; bootstrap support > 70% are shown. Note that the AN12‐1 species sequence is 100% identical to another reported AUS I sequence (GU086328); the characterised MEAM1 species sequence is 100% identical to other MEAM1 sequences reported from countries including the USA (GU086340, HM070411), Taiwan (GU086342), Egypt (DQ133373), Japan (AB204577) and Brazil (JN689356).
