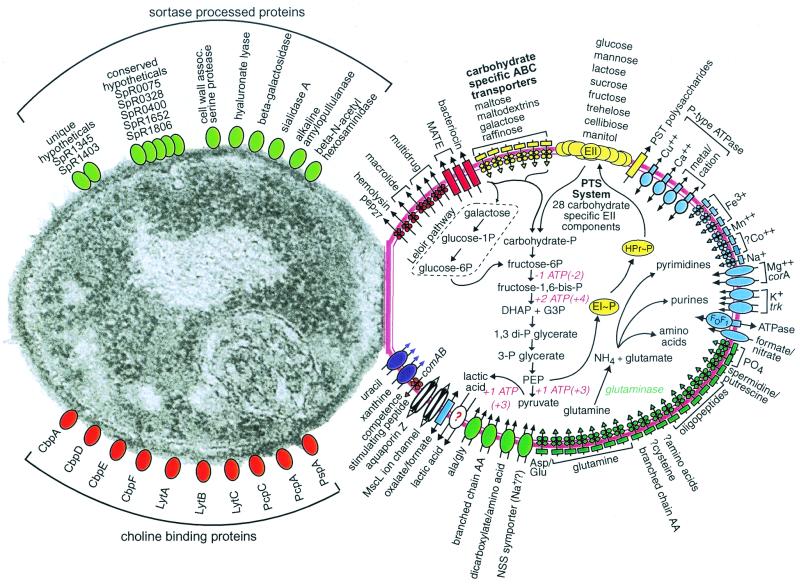
FIG. 1.
S. pneumoniae substrate transport, carbohydrate and glutamine metabolism, and selected categories of cell surface proteins. Transporters are shown in the cell on the right and are grouped by substrate specificity: multidrug and peptide exporters (red), carbohydrates (yellow), cations (blue), anions and amino acids (green), nucleosides, purines, and pyrimidines (purple), and other substrates (white). Question marks indicate uncertainty in the identity of the genes, direction of transport, or substrate. Permeases are drawn as rectangles; porins are drawn as cylinders; ATPases are drawn as ovals overlapping rectangles; other transporters are drawn as ovals; and ABC transporter elements are depicted as circles for nucleotide-binding proteins, diamonds for membrane-spanning permeases, and rectangles for the substrate-binding proteins. The 21 hypothetical ABC transporters for which no substrate could be predicted are not shown in the figure. Glutaminase, an enzyme we expected to find but could not identify, is listed in green italics. The glycolytic pathway leading to lactate is shown along with the pathway for glutamine to nitrogen-containing compounds. (Additional material on carbohydrate metabolism is at http://www.lilly.com/s.pneumoniae.) The ATP consumption and production values listed are for monosaccharides; ATP values for disaccharide catabolism are in parentheses. The choline-binding proteins are autolysin (LytA), endo-beta-N-acetylglucosaminidase (LytB), a surface protein involved in adherence and immunoglobulin A inactivation (CbpA), a putative lactoferrin-binding protein (PspA), and several surface proteins of unknown function (CbpD, PcpA, PcpC, PcpC1). Micrograph reprinted from reference 47 with permission of the publisher.