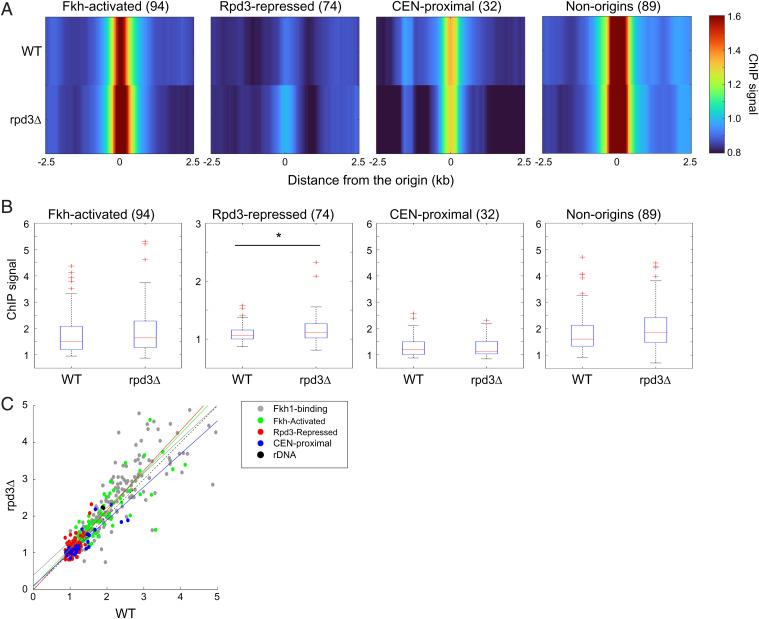
Fig. 5.
Rpd3 modulates Fkh1 binding to origins. Strains OAy1112 (FKH1-3xFLAG) and YHy17 (rpd3Δ, FKH1-3xFLAG) were synchronized in G1 phase and subjected to ChIP-seq. (A) Heatmaps of ChIP-seq enrichment at selected origin and Fkh1 binding loci; n of each group is indicated within parentheses. The “non-origins” group refers to Fkh1-binding sites called as peaks by MACS in the Fkh1 ChIP-seq dataset that do not overlap with a replication origin. (B) Box plots distributions of ChIP-seq enrichments for 500-bp regions aligned on origins of the indicated subgroups; n of origins or Fkh1 binding loci in each group is indicated within parentheses. Two-sided t tests were performed on all pairs of strains, and results are indicated as *P < 0.05 and **P < 0.01. (C) 2D scatter plots of ChIP-seq signals (500-bp regions) for 174 Fkh1-binding sites (called as peaks by MACS as described above but including origins) plus the indicated origin subgroups, color coded as indicated. A linear regression deriving the best-fit line is shown for each origin group.