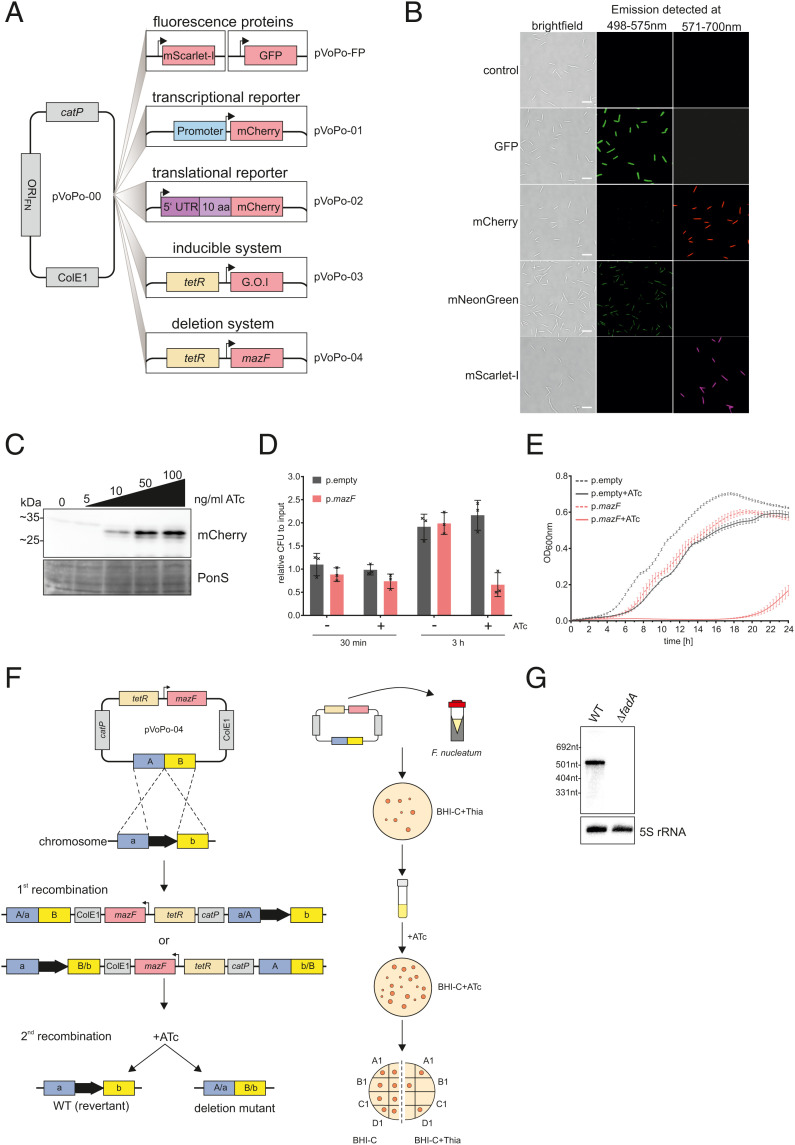
Fig. 2.
A genetic toolbox for F. nucleatum. (A) Overview of the plasmid-based genetic tools developed for F. nucleatum based on the vector pVoPo-00. catP, chloramphenicol resistance cassette; ColE1, replication of origin; FP, fluorescent protein (GFP, mCherry, mNeonGreen, mScarlet-I); G.O.I, gene of interest; ORIFN, origin of replication for F. nucleatum; tetR, tetracycline repressor. (B) Representative images of F. nucleatum carrying pVoPo-FP expressing different fluorescent proteins. The cells were PFA-fixated prior to overnight maturation at 4 °C. The emission was detected at the indicated wavelength. (Scale bars, 5 µm.) (C) Western blot analysis of lysates of F. nucleatum carrying a plasmid with mCherry introduced in the inducible system (pVoPo-03) after 30-min exposure to different ATc concentrations. mCherry runs as a duplet likely representing the full-length protein as well as a truncated version arising from an internal translational start site (53). Ponceau S (PonS) staining is shown as loading control. (D) Quantification of colony-forming units (CFU) for F. nucleatum carrying the empty vector control pVoPo-03 (p.empty) or the pVoPo-03-mazF plasmid (p.mazF). The bacteria were grown to mid-exponential phase and treated with 100 ng mL−1 ATc to induce mazF expression. Serial dilutions of the samples were plated after 0 min (input), 30 min, and 3 h. Untreated samples were used as control. Data are presented as the average and SD for three biological replicates relative to the input CFUs. (E) Growth curves for F. nucleatum carrying either p.empty or p.mazF in the presence or absence of 100 ng mL−1 ATc. No selection pressure for plasmid maintenance was included. Displayed is the average of three biological replicates with SD. (F) Schematic representation of allelic exchange (Left) and experimental workflow (Right) using the pVoPo-04 system to generate unmarked deletion strains. A/B, up- and downstream homology regions. (G) Northern blot detection of fadA using total RNA samples extracted from F. nucleatum WT or a ΔfadA strain generated via the deletion system pVoPo-04. 5S rRNA served as loading control.