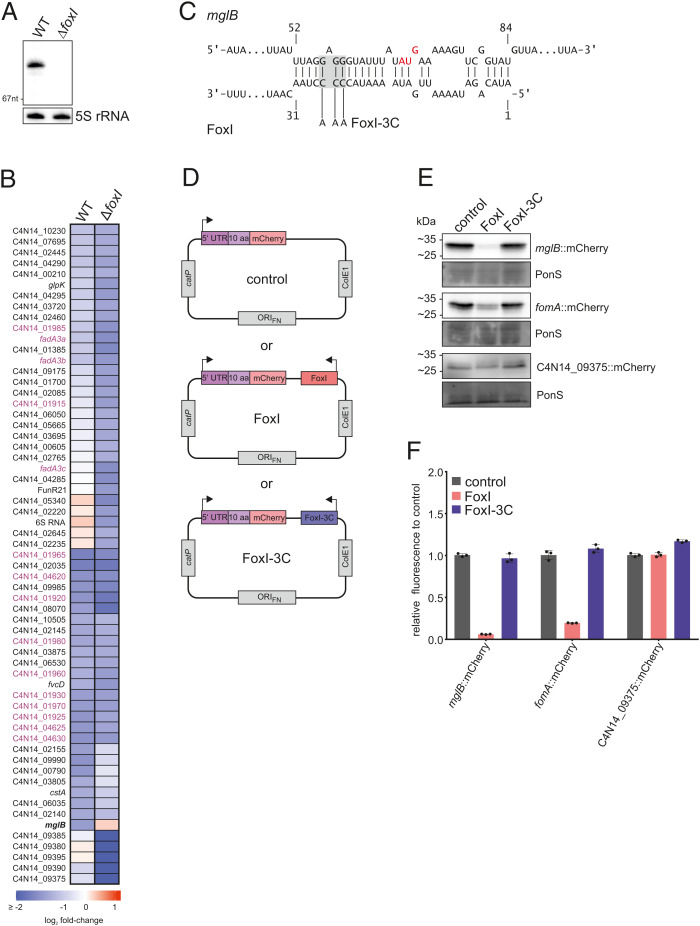
Fig. 5.
The sRNA FoxI as a negative regulator of the σE response. (A) Northern blot detection of FoxI using total RNA samples extracted from F. nucleatum WT or ΔfoxI generated via the deletion system pVoPo-04. The 5S rRNA served as loading control. (B) Differential gene expression upon σE induction in WT F. nucleatum or in the FoxI deletion strain (ΔfoxI). The heatmap displays log2 fold-changes of genes that are significantly down-regulated in either background (log2 fold-change ≤ −1; FDR ≤ 0.05). mglB is marked in bold as the only gene that is not down-regulated in the ΔfoxI background upon σE induction. Members of the three multicistronic operons started by FadA-domain containing genes are marked in purple. (C) Schematic representation of IntaRNA (93,) prediction of base-pairing between mglB mRNA and FoxI. The AUG start codon of mglB is marked in red. The mutation of the sRNA for FoxI-3C is indicated in gray. (D) Schematic representation of the translational reporter constructs used in E. F. nucleatum was either transformed with pVoPo-02 plasmids carrying mCherry fused to the 5′-region of the target gene only (control), or in combination with the expression cassette for FoxI (FoxI) or the seed region mutant FoxI-3C. (E) Representative western blots for each gene tested in the translational reporter system. C4N14_09375 served as control gene as it does not harbor any predicted FoxI-binding site. PonS staining is shown as loading control. (F) Quantification of mCherry by flow cytometry for the same constructs as shown in E. The average of three biological replicates relative to that of the control (control) is displayed together with the SD.