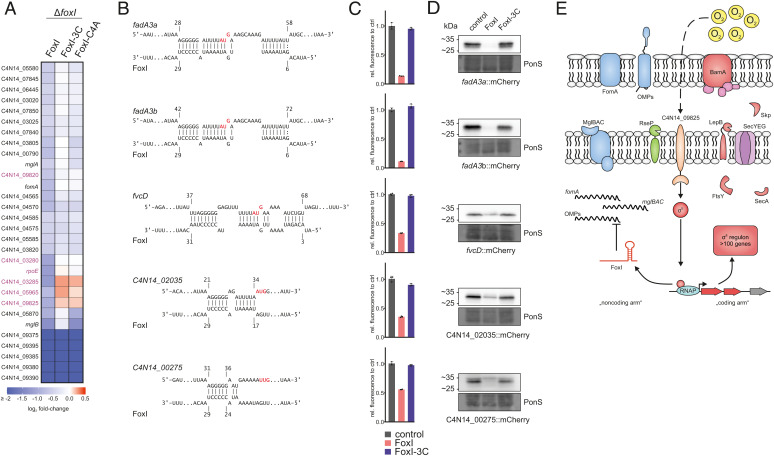
Fig. 6.
Extended target spectrum of the FoxI sRNA. (A) Overview of all significantly down-regulated genes (log2 fold-change ≤ −0.5; FDR ≤ 0.05) upon pulse expression of FoxI or the seed region mutants FoxI-3C or FoxI-C4A in the ΔfoxI background. The heatmap displays the log2 fold-changes. Genes of the identified σE regulon (Fig. 3) are marked in purple. (B) Schematic representation of IntaRNA target predictions between different target mRNAs and FoxI. The AUG start codons are marked in red. (C) Quantification of mCherry by flow cytometry for the translational reporter plasmids carrying the target 5′ region shown in B alone (control) or in the presence of FoxI or FoxI-3C. The average of three biological replicates relative to that of the control is displayed together with the SD. (D) Representative images of western blots of total protein samples for bacteria expressing the indicated reported constructs probed for mCherry. PonS staining is shown as loading control. (E) Model of the σE regulon in F. nucleatum. σE is released from its putative anti-σ factor (C4N14_09825) and up-regulates expression of its regulon consisting of >100 genes (marked in red). This includes bamA and skp, important for insertion of OMPs as well as lepB, ftsY, or secA, which are involved in protein-translocation across the IM. σE also activates the transcription of the sRNA FoxI. FoxI, in turn, down-regulates membrane-associated proteins such as the IM protein complex mglBAC and the OMP fomA as well as other OMPs.