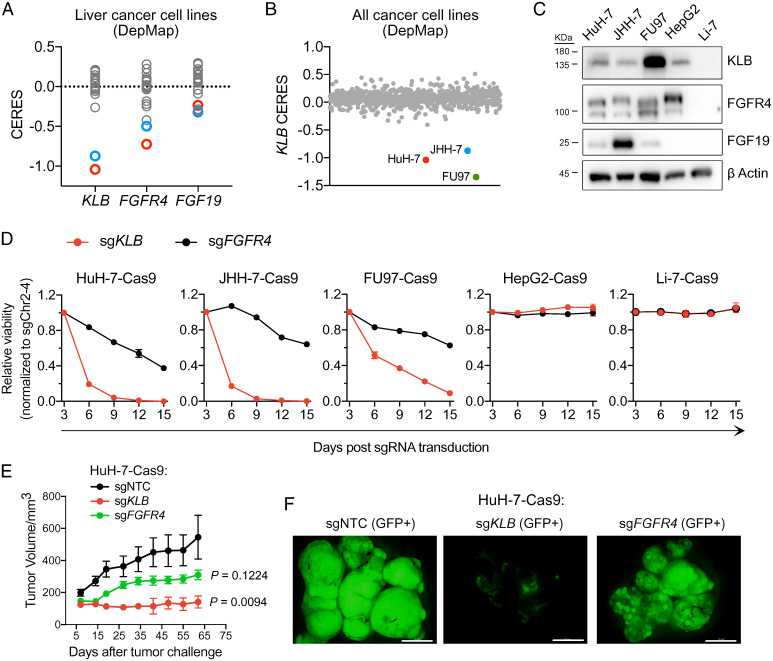
Fig. 2.
CRISPR inactivation of KLB displays a more severe fitness defect than that seen with inactivation of FGFR4 in FGF19-positive cancer cell lines. (A) CERES scores of KLB, FGFR4, and FGF19 in 22 liver cancer cell lines tested in CRISPR-Cas9 loss-of-viability screens (DepMap). (B) KLB CERES scores of 807 cell lines in DepMap. (C) Western blots showing expression levels of KLB, FGFR4, and FGF19 in indicated cell lines. (D) Viability effects after CRISPR inactivation of KLB or FGFR4 in indicated cell lines normalized to control (sgChr2-4). (E) Tumor growth curve after CRISPR inactivation of KLB or FGFR4 in HuH-7 cells xenografted subcutaneously into nude mice. Data are mean ± SEM, with n = 5 mice per group. Comparisons with the control group (sgNTC) were performed with ordinary one-way ANOVA (F [2, 12] = 5.904, P = 0.0164) followed by Dunnett multiple comparison test (sgKLB vs. sgNTC: P = 0.0094; sgFGFR4 vs. sgNTC: P = 0.1224). (F) Fluorescence images of dissected tumors on day 62 of E (scale bar, 1 cm).