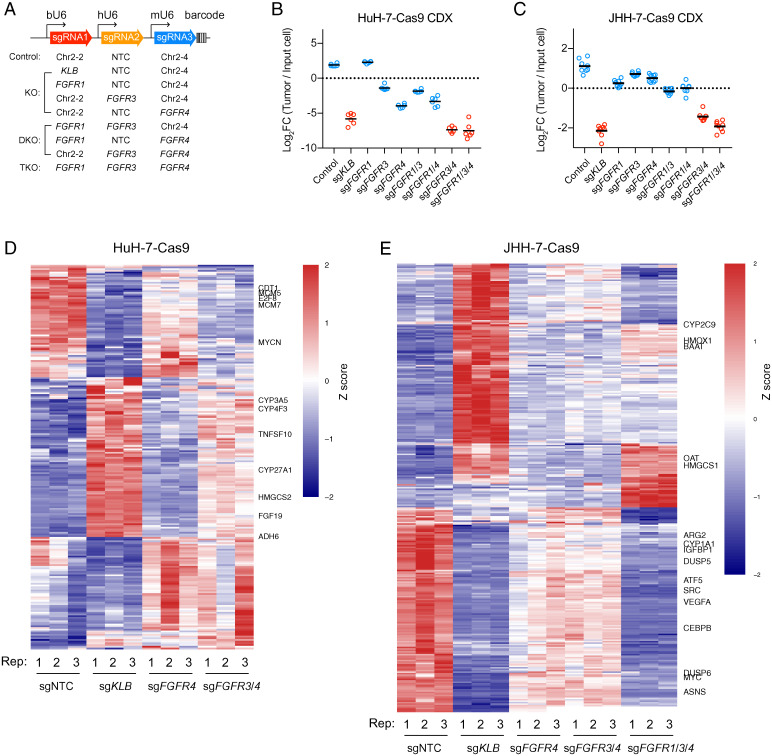
Fig. 5.
FGFR redundancy in HuH-7 and JHH-7 revealed by in vivo tumor growth and transcriptome analyses. (A) Barcoded polycistronic vectors expressing indicated combinations of sgRNAs driven by bovine, human, and mouse U6 promoters. (B and C) Relative abundance (log2 transformed) of HuH-7–Cas9 and JHH-7–Cas9 cells harboring indicated combinations of sgRNAs from subcutaneous cell line–derived tumor xenografts (CDX) normalized to input cells. Bars represent averages from n = 6 mice (HuH-7–Cas9) or n = 7 mice (JHH-7–Cas9). Comparisons among groups were performed with ordinary one-way ANOVA (FHuH-7 [8, 45] = 247.5; FJHH-7 [8, 63] = 206; P < 0.000001) followed by Tukey multiple comparison test (P values in SI Appendix, Tables S1 and S2). (D and E) Heat map showing differentially expressed genes in HuH-7–Cas9 and JHH-7–Cas9 cells after CRISPR inactivation of indicated genes relative to control (sgNTC). The genes were filtered as described in Materials and Methods. Row-wise z score transformation was performed. DKO, double knockout; FC, fold change; KO, knockout; TKO, triple knockout.