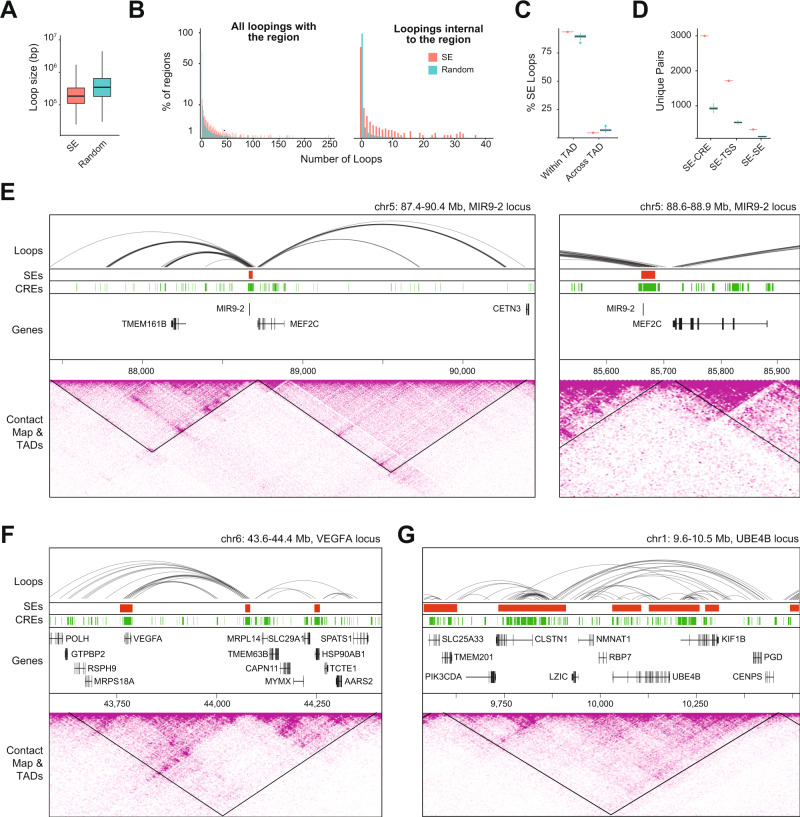
Fig. 4. SEs display a distinct chromatin looping pattern.
A Size distribution of loops intersecting with at least one SE (n = 14,491 loops) or random SE-sized genomic region (n = 299,795 total loops across 100 randomly generated regions, see methods). Boxplots represent the median and interquartile range (IQR); whiskers mark 1.5x the IQR; data beyond 1.5x the IQR are plotted as individual points. B Distribution of loops per SE or random SE-sized genomic region either making contact with at least one foot touching an SE/random region (left) or both feet contained within a single SE/random region (right). C Percentage of loops in contact with SEs (n = 1) or random regions (n = 100) that cross TAD boundaries. Boxplots represent the median and interquartile range (IQR); whiskers mark 1.5x the IQR; data beyond 1.5x the IQR are plotted as individual points. D Number of unique pairs of features connected by chromatin looping (n = 1 SEs dataset, n = 100 random regions datasets). Boxplots represent the median and interquartile range (IQR); whiskers mark 1.5x the IQR; data beyond 1.5x the IQR are plotted as individual points. E–G Chromatin loops, SEs, CREs, TADs, and Hi-C contact maps for the [E] MIR9-2 (zoom-in on right panel), [F] VEGFA, and [G] UBE4B loci. Abbreviations SE Super-enhancer, TAD Topologically associating domain, CRE Cis-regulatory element, TSS Transcription start site.