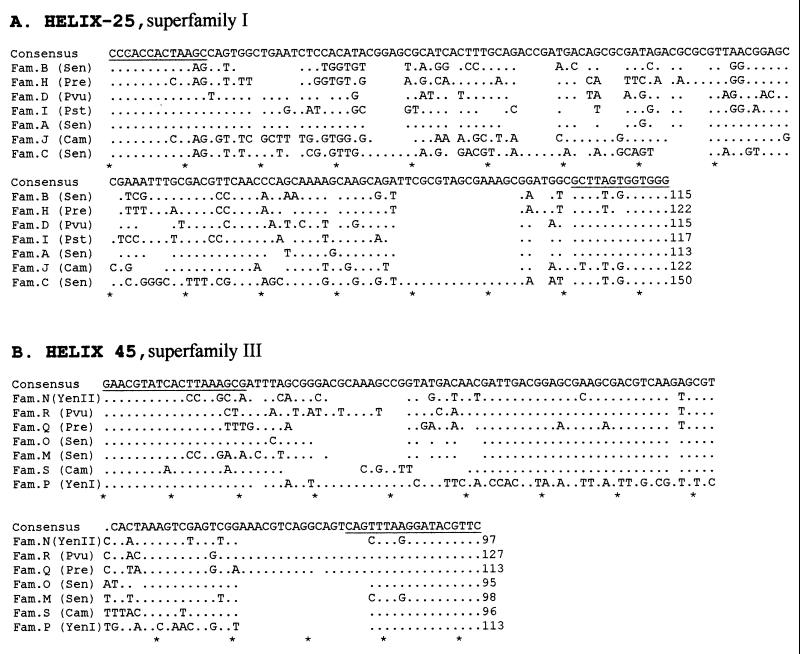
FIG. 4.
Alignment of IVSs using CLUSTAL X version 1.6 for the Macintosh. The alignment was imported into and modified in Word 98. Conserved bases are shown by dots, and deletions are shown by spaces. The consensus sequence is shown at the top of the alignment, and each 10th base is marked by an asterisk at the bottom. The number of bases on the IVS is at the end of each sequence. The bases which are underlined are postulated to be part of the primary stem, as described in the stem-loop structure postulated for the IVS based on secondary structure (22, 33). (A) Helix 25 IVSs from S. enterica (Sen) (families A, B, and C), Proteus vulgaris (Pvu) (family D), P. rettgeri (Pre) (family H), P. stuartii (Pst) (family I), and C. amalonaticus (Cam) and K. oxytoca (Kox) (family J). (B) Helix 45 IVSs from S. enterica (Sen) (families M and O), Yersinia enterocolitica (Yen) (families N and P), Proteus vulgaris (Pvu) (family R), P. rettgeri (Pre) (family Q), and C. amalonaticus (Cam) (family S).