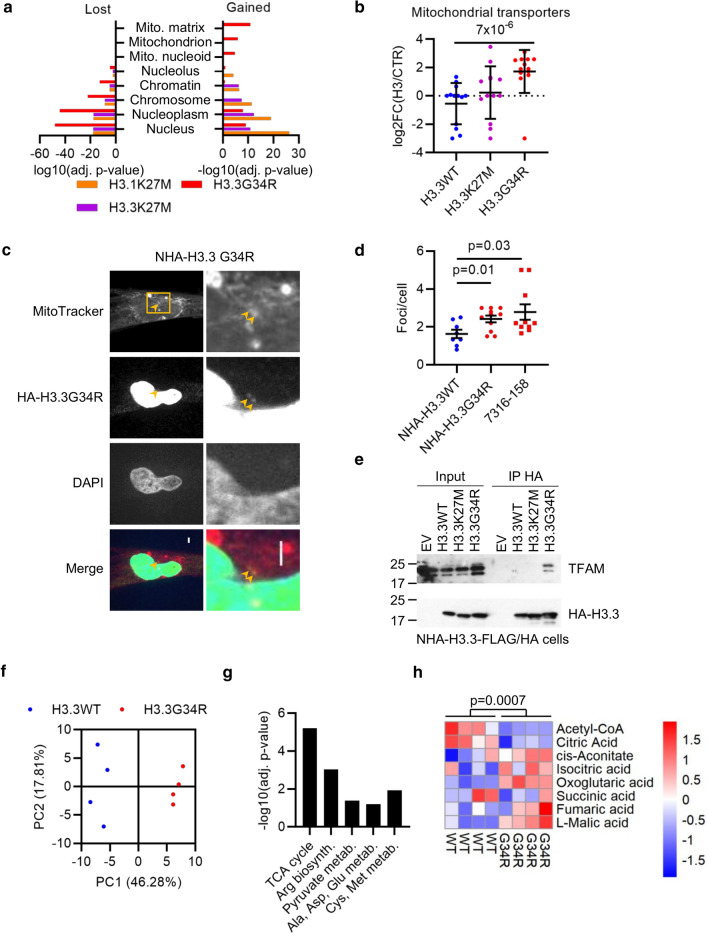
Fig. 2.
Mitochondrial localization of H3.3G34R. A Enrichment of subcellular localizations (Gene Ontology Cellular Compartments) among the proteins differentially bound by each oncohistone. B Relative enrichment [log2(H3/control)] of outer (TOMM) and inner (TIMM) mitochondrial transporter proteins associated with each histone. p: t test. C Confocal microscopy of NHA cells expressing H3.3G34R-FLAG/HA and stained with MitoTracker Red and HA. Box shows zoom area at right. Arrows mark co-localization of H3.3G34R with mitochondria. Scale bar: 3 μm. D Quantification of colocalization between MitoTracker Red and H3.3 in NHA cells expressing H3.3WT or H3.3G34R (antibody: HA tag) or 7316-158 cells (antibody: H3.3G34R). Bars show mean ± standard error of at least 7 fields of view. E Co-immunoprecipitation of H3-FLAG/HA and TFAM from NHA cells transduced with indicated constructs and subjected to HA IP. F Principal components analysis of normalized metabolite concentrations in NHAs expressing H3.3WT or H3.3G34R as determined by LC–MS (n = 4). G Enriched metabolic pathways in H3.3G34R vs H3.3WT NHAs. H Relative concentration of indicated TCA metabolites in H3-expressing NHA cell lines (n = 4). p: t test