Figure 6. Transcriptomic and T-cell receptor (TCR) sequencing of cultured tumor-infiltrating lymphocytes (TIL) shows preferential expansion of specific TIL states.
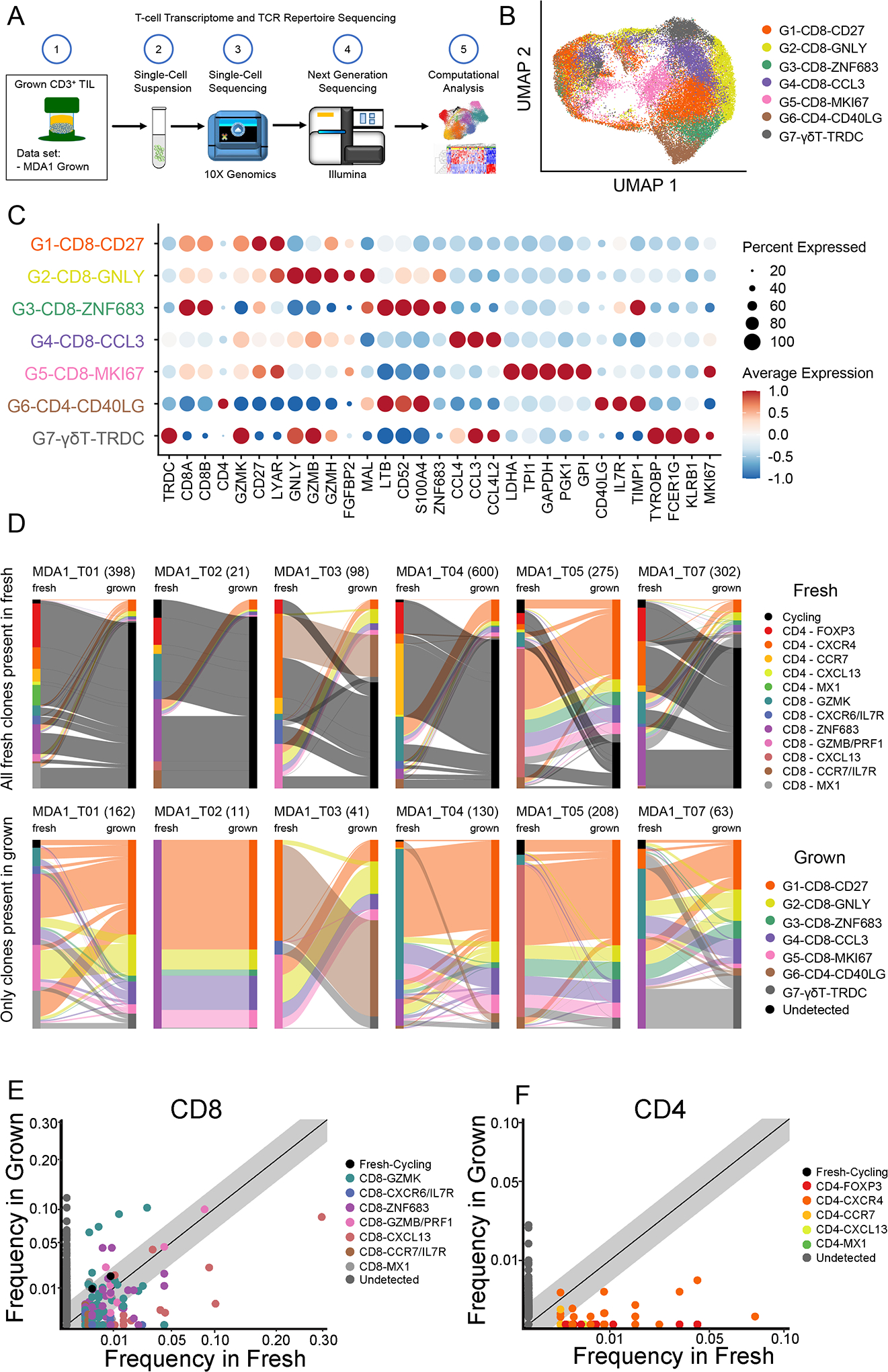
(A) From the same samples that make up the MDA1 data set, tissue was also used to generate TIL. The scheme is shown, in which TIL grown from these samples were sent for single-cell RNA sequencing to generate transcriptomic and TCR analyses. (B) The uniform manifold approximation and projection (UMAP) projection of 40,605 single T cells from six pancreatic ductal adenocarcinoma (PDAC) TIL cultures, showing five CD8, one CD4, and one γδ T-cell cell states. Each dot is a single cell and is colored according to transcriptomic state. (C) Top differentially expressed genes for each cell state are shown. Expression is marked by a circle, where greater size indicates a greater percentage of cells expressing this gene and the color gradient indicates a higher level of expression as it moves from blue to red. (D) Sankey plot visualization shows the transcriptomic cell state in which the TCR initially started in the tissue and where it finished in the culture (fresh tissue on left and grown TIL on right). Clonotypes are classified by the cell state the majority of that clonotype is found. For each sample/graph, two plots are shown with the upper row of graphs showing all clones detected in the fresh tissue and the bottom row showing only those clones detected in both the fresh tissue and grown TIL. Total number of clones are shown in parentheses. Scatter plot for (E) CD8+ TIL and (F) CD4+ TIL comparing the relative frequency of T-cell clones in the fresh pancreatic ductal adenocarcinoma (PDAC) tissue versus the grown TIL. Each dot is a single clonotype and is colored by the transcriptomic state from the fresh TIL. Clonotypes that were not detected in both the fresh tissue and grown culture are colored gray. The diagonal line indicates a 1:1 relative frequency, where clones above the line are found at greater relative frequency in the grown than in the fresh tissue and vice versa.
