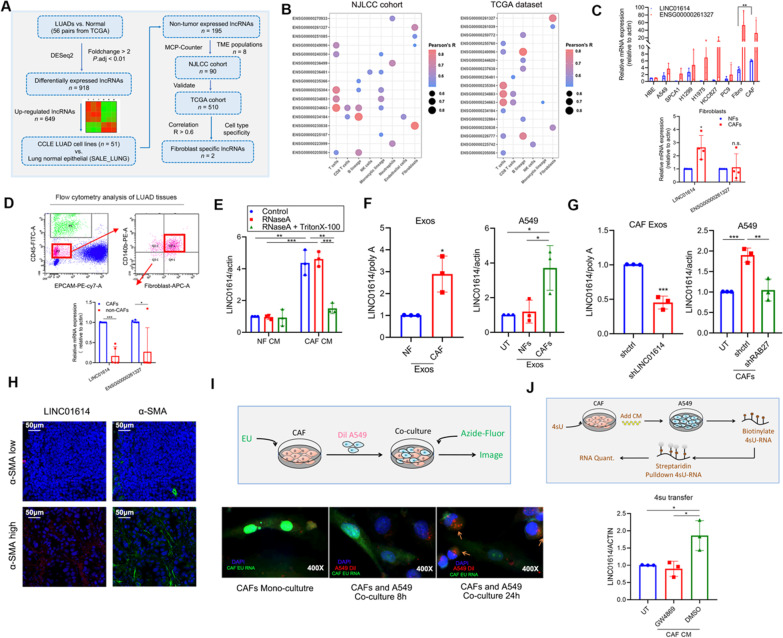
Fig. 2.
Intercellular transfer of a CAF-specific lncRNA LINC01614 by exosomes. A Schematic showing the screening strategy of TME-specific lncRNAs. B LncRNAs that are moderately to highly positively correlated with TME cells in the NJLCC cohort (left) and TCGA database (right) (ENSG0000230838 is for LINC01614). C qRT-PCR of LINC01614 (ENSG0000230838) and ENSG00000261327 in CAFs, NFs, MRC5, and LUAD cell lines (n = 3). D Flow cytometric sorting of CAFs from cell suspension of human LUAD tissue. Live/Dead Fixable Viability Dye staining was used to detect dead cells. CD45 was used as an immune cell marker, EpCAM as an epithelial cell marker, fibroblast-specific protein (FSP) and PDGFR β (CD140b) as fibroblast markers. qRT-PCR analysis of LINC01614 and ENSG00000261327 in isolated CAFs and CAF-depleted cells (n = 3). E qRT-PCR of LINC01614 and ENSG00000261327 in the CM of A549 cells treated with RNase (2 μg mL−1) alone or combined with Triton-X-100 (0.1%) for 20 min (n = 3). F qRT-PCR analysis of LINC01614 in CAFs exosomes versus NFs exosomes (n = 3), and A549 cells with indicated treatments versus those cultured alone for 24 h (n = 3). G Relative expressions of LINC01614 in CAF-derived exosomes (left) and the A549 cells co-cultured with the CAFs transfected with shRAB27 (right) were determined by qRT-PCR. H Representative images of α-SMA immunofluorescent staining and LINC01614 fluorescence in situ hybridization staining in LUAD patient tissues (n = 10). I Schema and representative images for measuring RNA transfer from CAFs to A549 cells utilizing the uridine analog EU for fluorescence microscopy (green). CAFs were labeled with EU and co-cultured with Dil lipid-labeled A549 cells for the indicated time. Representative images of EU-positive A549 cells (orange) are shown (n = 3). J Schema for measuring LINC01614 transfer from CAFs to A549 cells. Conditioned medium from CAFs labeled with 4sU was added to A549 cells, 4sU RNA in A549 cells was isolated with streptavidin pull-down. Relative transfer of 4sU RNA to mono-cultured A549 cells the addition of CM collected from 4sU-labeled CAFs. CAFs CM depleted of exosomes with GW4869 is shown as a control for exosome-dependency (n = 3). For all experiments, means ± s.d. are shown, and independent sample t-tests determined P values. *P < 0.05, **P < 0.01, ***P < 0.001. TME tumor microenvironment; NJLCC Nanjing lung cancer cohort; UT cancer cells without any treatment; CM conditioned medium; Exos, exosomes; EU 5-ethynyl uridine; 4sU 4-thiouridine; LUAD lung adenocarcinoma.
Source data are provided