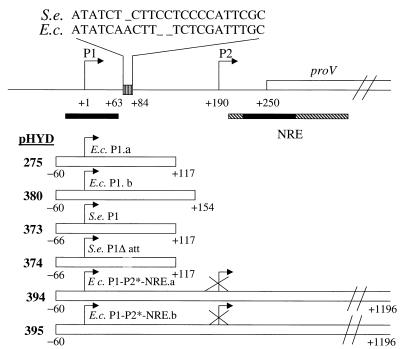
FIG. 1.
Description of plasmids used for studies of in vivo expression from the proU P1 promoter. The line on top depicts the proximal region of the proU operon, and the positions of the cis regulatory elements (namely, P1, P2, and the NRE) are marked relative to that of its first structural gene, proV. Nucleotide number designations are given, with the start site of P1 transcription taken as +1. Solid bars, in the vicinity of P1 and within the NRE, mark the positions of bent-DNA motifs in this region. The box with vertical stripes depicts the location of a 22-base C-rich segment on the P1-initiated transcript of S. enterica (S.e.). The sequence of this segment from S. enterica and from E. coli (E.c.) is shown above the line. Beneath are represented (by the individual open bars) the extents of DNA from the proU regulatory region carried upstream of the lacZ reporter gene in the single-copy-number plasmids (whose pHYD number designations, and abbreviated descriptions, are given alongside) that were used for the in vivo expression studies. Deletion of the 22-base region in pHYD374 is denoted by the interrupted line segment. Note that the P2 promoter in each of the plasmids pHYD394 and pHYD395 has been mutationally inactivated.