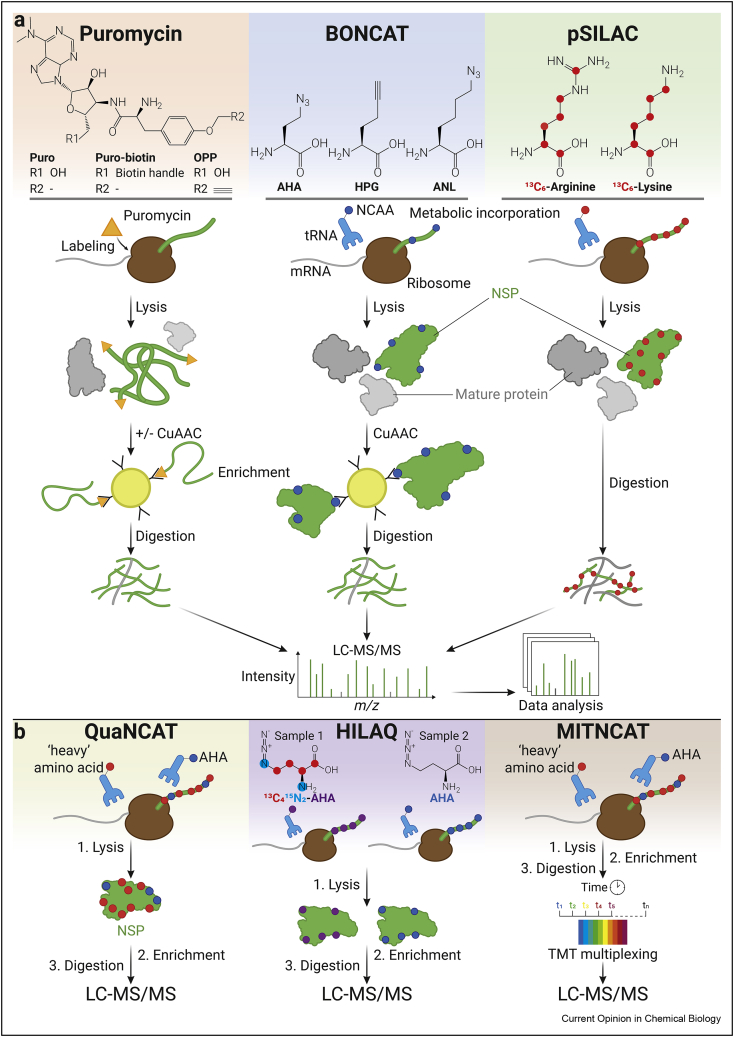
Figure 2.
Mass spectrometry–based methods for analysis of newly synthesized proteins. (a) Currently used MS-based methods to analyze NSPs can be categorized into three main strategies. 1. The puromycin-based strategy relies on the aminonucleoside antibiotic, puromycin (Puro), which inhibits protein synthesis and couples to the C-terminus of nascent polypeptide chains (NPCs). Biotinylated and alkynylated variants of puromycin enable targeted enrichment and measurement of NPCs by LC-MS; 2. BONCAT-based methods rely on the methionine surrogates AHA, HPG, or ANL to enrich for NSPs. After their metabolic incorporation in NSPs, a copper (I)-catalyzed alkyne–azide cycloaddition (CuAAC) can be used to functionalize labeled NSPs with affinity handles, such as biotin, to enable enrichment; 3. SILAC relies on metabolic labeling of NSPs with isotopically labeled amino acids. The strategy does not contain an enrichment step, but NSPs can be identified by LC-MS/MS detection of the isotopically labeled amino acids. (b) Combinations of BONCAT with other quantitative techniques to enhance the accuracy and temporal resolution of NSP analysis. QuaNCAT combines BONCAT with pSILAC, to discriminate between bona fide NSPs and false positives. Heavy isotope-labeled AHA quantification (HILAQ) uses stable isotope-labeled AHA and nonlabeled AHA for relative quantification of two different conditions. MITNCAT (multiplex isobaric tagging/noncanonical amino acid tagging) combines QuaNCAT and TMT multiplexing to reduce labeling time and allows detection of small changes in protein synthesis in short timeframes.