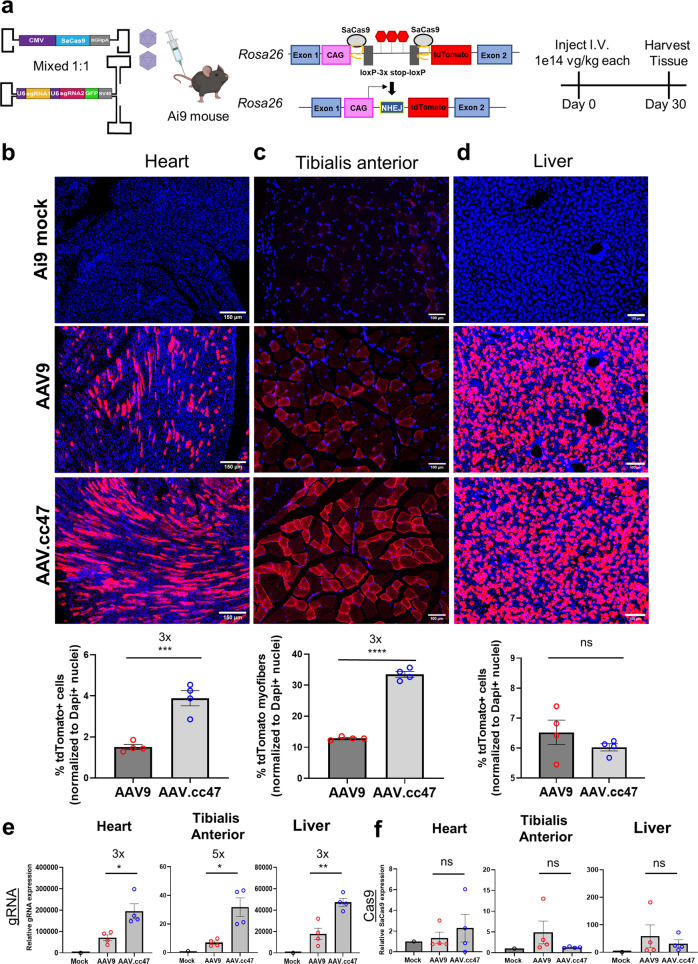
Fig. 4. Systemic administration of AAV.cc47-CRISPR increases genome editing efficiencies in heart and skeletal muscle of Ai9 reporter mice compared to AAV9.
a 8–10 week old Ai9 reporter mice (n = 4) were injected with AAV.cc47 (blue) or AAV9 (red) using our lead dual AAV vector system consisting of a single-stranded genome with cytomegalovirus (CMV) driven SaCas9 and a self-complementary genome with 2 gRNAs driven by U6 promoters targeting the Rosa26 locus. The total dose administered was 2.0e14 vg/kg (1.0e14 vg/kg each vector). Representative immunofluorescence images for tdTomato (red) and histological quantification in heart (b), skeletal muscle (c), and liver (d). Fifty micrometers thick cross sections were obtained via a vibratome for the heart and liver, while 7 μm thick cross sections were obtained for skeletal muscle via cryostat. Quantification of the percentage of tdTomato+ cells (heart and liver) and myofibers (tibialis anterior) was normalized to the number of DAPI + nuclei in each tissue. For all histological quantification, 2 sections per mouse and 3 images per section (total of 6 images) were used to quantify % transduced cells and myofibers using ImageJ. Quantitative RT-PCR was performed with primers amplifying each gRNA (e) or SaCas9 (f) mRNA relative to mouse Actb. Each dot represents an individual mouse, fold change is listed above significance, the bar graphs represent the mean value and error bars represent the standard error mean. Statistical significance was determined by a two-tailed Student t Test (2 treatment groups; heart, P < 0.0009; tibialis anterior, P < 0.0227; liver, P < 0.2892) or One-Way ANOVA with Tukey’s posttest (3 treatment groups; heart gRNA, P < 0.0314, Cas9, P < 0.7966; tibialis anterior gRNA, P < 0.0348, Cas9, P < 0.4459; liver gRNA, P < 0.0126, Cas9, P < 0.8205). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns not significant. Source data are provided as a Source Data file.