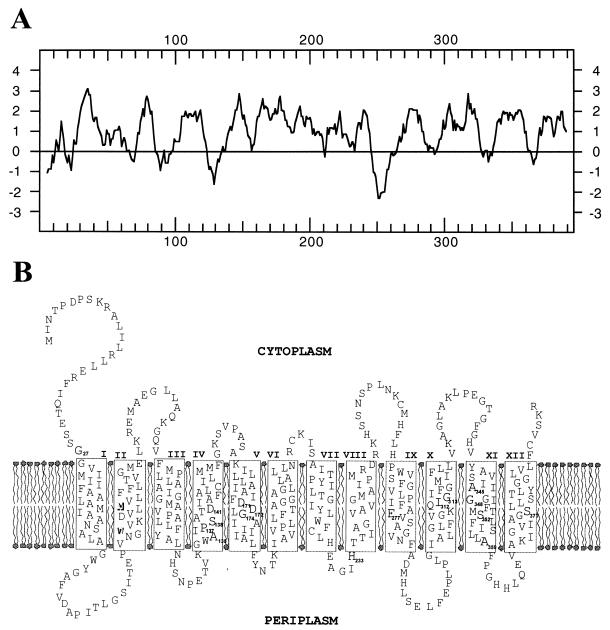
FIG. 3.
Hydropathy plot (A) and model for the secondary structure (B) of NhaA encoded by pAM2. (A) Hydropathy analysis of NhaA was performed according to the method of Kyte and Doolittle (20). Positive values represent high hydrophobicity, and negative values indicate low hydrophobicity, averaged over a window of seven amino acids. (B) The model of secondary structure for NhaA is based on the hydropathy analysis of the deduced gene product, a transmembrane segment prediction using the HMMTOP computer program (43) and the model reported for NhaA of E. coli (27, 39). Putative transmembrane segments are shown in boxes connected by hydrophilic segments. The Roman numerals indicate the numbers of the transmembrane segments.