Summary
Interactions between angiogenesis and neurogenesis regulate embryonic brain development. However, a comprehensive understanding of the stages of vascular cell maturation is lacking, especially in the prenatal human brain. Using fluorescence-activated cell sorting, single-cell transcriptomics, and histological and ultrastructural analyses, we show that an ensemble of endothelial and mural cell subtypes tile the brain vasculature during the second trimester. These vascular cells follow distinct developmental trajectories and utilize diverse signaling mechanisms, including Collagen, Laminin, and Midkine, to facilitate cell-cell communication and maturation. Interestingly, our results reveal that tip cells, a subtype of endothelial cells, are highly enriched near the ventricular zone, the site of active neurogenesis. Consistent with these observations, prenatal vascular cells transplanted into cortical organoids exhibit restricted lineage potential that favors tip cells, promote neurogenesis, and reduce cellular stress. Together, our results uncover important mechanisms into vascular maturation during this critical period of human brain development.
Graphical Abstract

In brief
The transcriptomic and functional properties of diverse endothelial and mural cells in the nascent vasculature in second trimester human brain are revealed, shedding light on vascular trajectories and heterogeneity in human brain development.
Introduction
The vasculature is increasingly recognized to impact brain function in health and disease (Licht and Keshet, 2015; Silva-Vargas et al., 2013). Endothelial and mural cells constitute the main structural and functional vascular elements. Endothelial cells form the lumen of blood vessels and also secrete factors that influence neurogenesis in the mouse brain (Silva-Vargas et al., 2013). Mural cells, which include smooth muscle cells and pericytes, are located outside the endothelial cells. They are critical for the development of the blood-brain barrier (BBB) and neurovascular coupling (Armulik et al., 2010; Daneman et al., 2010; Hall et al., 2014; Kisler et al., 2017; Lacar et al., 2012). Brain endothelial cells are thought to originate from a perineural vascular plexus external to the brain parenchyma (Hogan et al., 2004; James et al., 2009), whereas the ontogeny of mural cells remains elusive (Armulik et al., 2011; Yamamoto et al., 2017). While recent results have shed light on the contributions of vascular cells to brain aging and neurodegeneration (Garcia et al., 2022; Winkler et al., 2022; Yang et al., 2022), a comprehensive understanding of the stages of vascular cell development in the prenatal brain is lacking.
In the developing mouse cortex, the nascent vasculature establishes distinct regional characteristics; the neural progenitors in the ventricular zone exist in a hypoxic avascular niche while oligodendrocyte precursor cells and basal progenitors prefer a perivascular location (Komabayashi-Suzuki et al., 2019). In subcortical regions, physical contact between vasculature and neural progenitors regulates key aspects of differentiation. Vascular filopodia in the ganglionic eminences contact radial glia, which elongates their cell cycle and favors neuronal differentiation (Di Marco et al., 2020). Radial glia also extend fibers which are anchored to local blood vessels; disruption of this interaction decreases the number of interneurons produced (Tan et al., 2016).
In the adult mouse brain, single-cell RNA sequencing (scRNA-seq) reveals three distinct subtypes of endothelial cells, including venous, capillary and arterial subtypes (Vanlandewijck et al., 2018). Compared to the adult mouse brain, the subtypes of endothelial cells in postnatal day 7 (P7) mouse brain expand to include mitotic and tip cells (Sabbagh et al., 2018). Trajectory analysis of P7 mouse endothelial cells suggests the presence of a hierarchical structure in which progenitor-like venous and mitotic cells give rise to arterial and tip cell phenotypes (Sabbagh et al., 2018). Similar to endothelial cells, scRNA-seq data identifies several mural cell subtypes in adult mouse brain, including smooth muscle cells, classic pericytes, and parenchymal fibroblasts (Vanlandewijck et al., 2018). While these studies revealed brain vascular transcriptomics in mice, the developmental origins and progression of vascular cells in the prenatal human brain remain unknown.
Results
Angiogenesis in the Prenatal Human Brain
To study angiogenesis in the prenatal human brain, we used a CD31 antibody to label endothelial cells, and NG2 and PDGFR-β antibodies to identify mural cells in the pallium, including the cerebral cortex, the ventricular zone/subventricular zone (VZ/SVZ) regions, and the subpallium, including the ganglionic eminences (GE) from 14–39 gestational weeks (GW) (Figure 1A–B, Figure S1A). Given the presence of active neurogenesis in VZ/SVZ in the pallium and the GE in the subpallium, we divided these regions into three zones based on the distance to the ventricle. Zone 1, defined by the expression of Nestin and PDGFR-β, included the VZ and SVZ up to 100 μm and contained abundant radial glia. Zone 2 contained densely packed immature neurons between 100 μm and 1 mm from the VZ/SVZ. Finally, Zone 3 defined 500 μm regions distal to Zone 2 (Figure 1C, Figure S1A–B). Quantification showed that CD31+ endothelial cell surface area was significantly higher in Zone 1 in the pallial and subpallial VZ/SVZ at 14–17 and 21–25 GW (Figure 1C–D, Figure S1B–C). At 39 GW, the CD31+ density in the pallial and subpallial VZ/SVZ regions showed no difference compared to those in Zones 2 and 3 (Figure 1D, Figure S1A, C). Similarly, NG2+ mural cell density was significantly higher in Zone 1 of the pallial and subpallial VZ/SVZ at 14–17 GW, but not at 21–25 GW (Figure 1C, E, Figure S1B, D). At 39 GW, NG2 and PDGFR-β antibodies detected non-vascular glial ribbons in VZ/SVZ and therefore were not quantified (Figure S1A). Unlike the results in the pallial and subpallial VZ/SVZ, endothelial cell density in the cortex showed uniform density similar to Zone 3 at all ages (Figure S1E–F). 3D reconstructions showed that many CD31+ endothelial cells at 14–17 GW contained abundant filopodia interdigitated with NG2+ mural cells in Zone 1, which were significantly reduced in Zones 2 and 3 (Figure 1F–G, Movie S1). The density of filopodia in Zone 1 reduced drastically at 21–25 GW (Figure 1G). Since filopodia are associated with active angiogenesis, we also quantified vascular branch points. These results showed that Zone 1 indeed contained more branch points than Zones 2–3 at 14–17 GW (Figure 1H). In addition, at 14–17 GW, Zone 1 contained more Ki-67+ endothelial (Ki-67+;CD31+) and mural (Ki-67+;PDGFR-β+) cells compared to Zone 3 (Figure 1I–K, Figure S1G–I, Movie S2). These findings indicate that the neurogenic ventricular zone in the prenatal human brain is also highly angiogenic.
Figure 1. Angiogenesis in the periventricular region of the germinal matrix in the prenatal human brain.
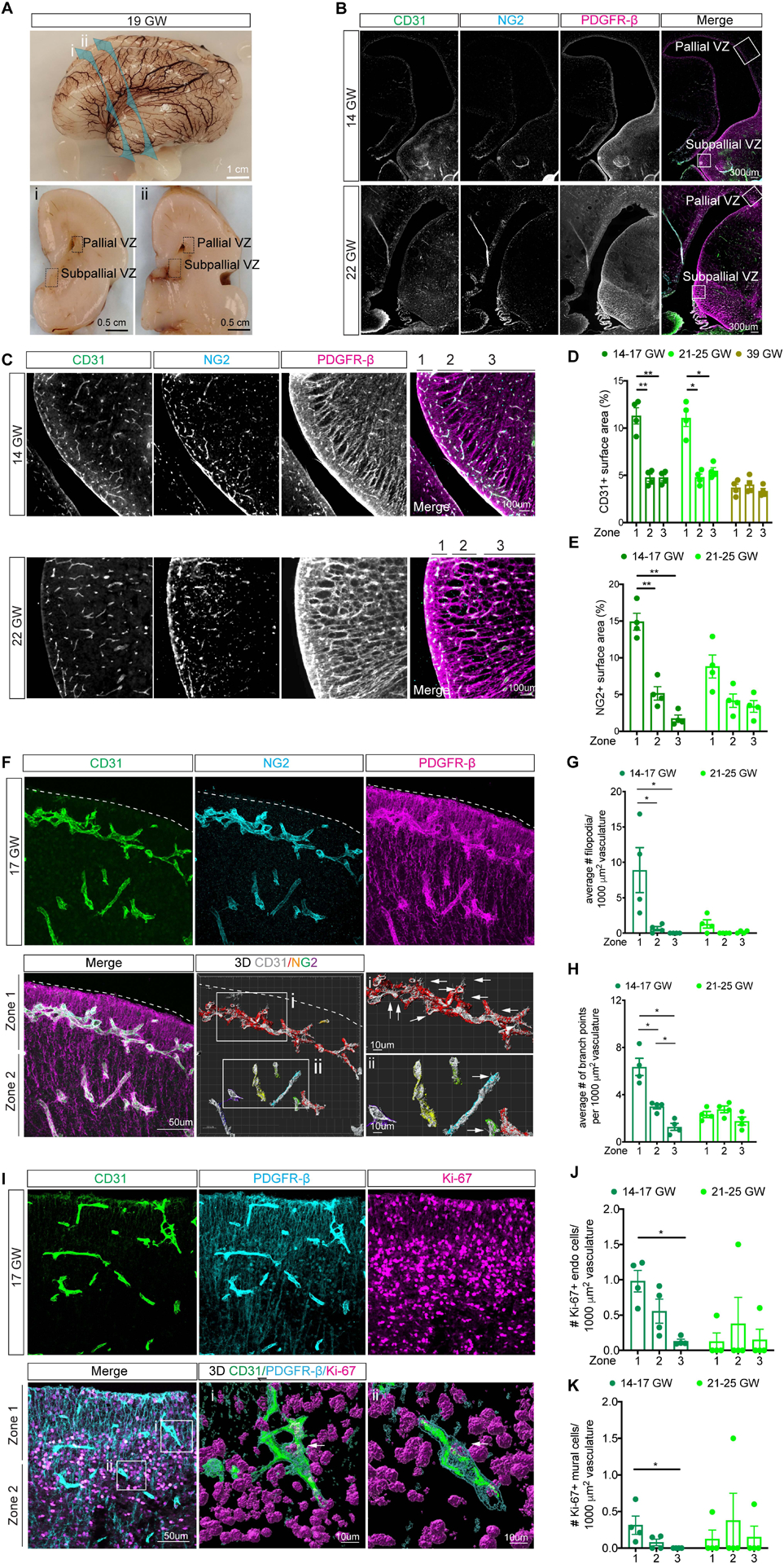
A. Gross photos of 19 GW prenatal human brain. Top: Lateral view showing the coronal planes for the bottom panels.
B. Confocal images of the periventricular area of human brain at 14 and 22 GW.
C. Immunostaining of CD31, NG2 and PDGFR-β in representative sections of the human medial ganglionic eminence (MGE) at 14 and 22 GW.
D-E. Quantification of CD31+ and NG2+ surface area in MGE.
F. Confocal image of 17 GW MGE immunostained for CD31, NG2 and PDGFR-β. 3D Imaris rendering shows CD31 (gray) and NG2 immunostaining with a gradient. Red indicates the highest NG2 expression to the lowest level in purple. The areas in the white box highlight filopodia (arrows).
G-H. Quantification of the number of filopodia (G) or branch points (H) in MGE.
I. Confocal image of the 17 GW MGE immunostained for CD31, Ki-67 and PDGFR-β. 3D images show Ki-67+ endothelial cell in i and Ki-67+ mural cell in ii.
J-K. Quantification of Ki-67+ endothelial (J) or mural (K) cells in MGE.
In D, E, G, H, J, and K, each data point represents the average of 5 sections from one case. Statistics used ANOVAs with post-hoc Student’s t-tests to assess differences within groups. Data are mean ± S.E.M. Non-significant comparisons not shown.
Ultrastructural Features of Prenatal Brain Vascular Cells
Next, we used transmission electron microscopy (TEM) to examine blood vessels in VZ and SVZ in GE from 17–23 GW (Figure 2A, magenta-shaded areas). At least three distinct populations of endothelial cells can be discerned based on their ultrastructural features (Figure 2, Figure S2, Table S1). The first endothelial cell subtype exhibited light translucent cytoplasm that contained few polyribosomes, rough endoplasmic reticulum (RER), small Golgi apparatus, and few caveolae and clathrin-coated vesicles (Figure 2B–J, M–T; Figure S2B–E, G–I, shaded in lavender). The second subtype contained dark, electron-dense cytoplasm and had abundant polyribosomes, RER, Golgi, and caveola-like vacuoles (Figure 2B–J, M–T; Figure S2B–E, G–I, shaded in green). The third subtype contained intermediate electron lucent cytoplasm with filopodia (Figure 2B–J, M–T; Figure S2B–E, G–I, shaded in blue). Dark endothelial cells had more mitochondrial mass at 17 GW compared to light endothelial cells and overall endothelial cells at 17 GW compared to 21 GW contained less mitochondrial mass (Figure 2P–U). In addition, all three endothelial cell subtypes displayed prominent RER, whose connection to mitochondria may contribute to angiogenesis (Wang et al., 2021). Other features characteristic of endothelial cells included junctional complexes (Figure 2B–G, Figure S2C, 2I, double arrows) and Weibel-Palade bodies (Figure 2H–I). The endothelial cells were covered by sparse and discontinuous basal lamina, indicative of immature BBB (Figure 2D, 2G, Figure S2D). Some blood vessels in the VZ/SVZ at 17 GW contained no detectable lumen, suggesting that these were nascent sprouts that were not yet perfused (Figure 2N, Figure S2C–D). By 23 GW, most vessels in these regions showed small lumens, some containing red blood cells (Figure S2A).
Figure 2. Ultrastructural features of developing human MGE and cortical vasculature.
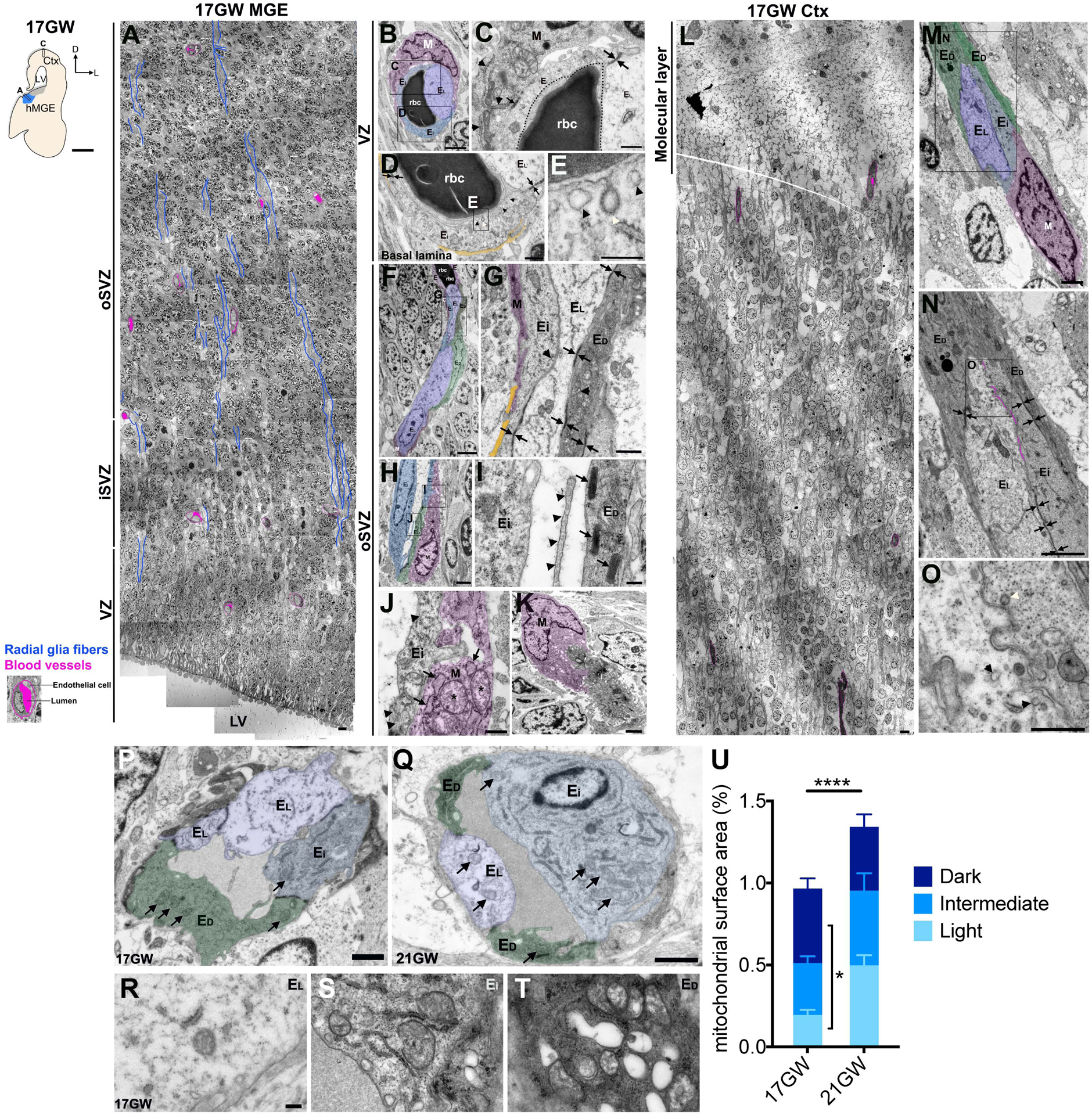
A. Composite images of ultrathin sections from 17 GW MGE. The magenta outlines the vasculature, the dense magenta spots highlight the lumen of each blood vessel, and blue shows radial glial fibers.
B-E. Blood vessel in the VZ composed of intermediate (blue) and light (purple) endothelial cells and a mural cell (magenta).
C. High magnification of cells in B showing the mural cell cytoplasm with abundant RER (arrowheads) and adherens junctions (arrow). Intermediate and light endothelial cells show tight junctions (arrows). A dotted black line indicates the blood vessel lumen.
D. Tight junctions (arrows) between endothelial cells and discontinuous basal lamina (yellow).
E. High magnification from B highlighting the caveolae (black arrowheads) and clathrin coated vesicles (white arrowhead).
F-G. Blood vessel in the oSVZ showing light (EL), intermediate (Ei) and dark (ED) endothelial cells and mural cell expansions (M, magenta). Note the long tight junctions (arrows) and discontinuous basal lamina (yellow).
H-I. A blood vessel composed of intermediate (blue) and dark (green) endothelial cells and a mural cell (magenta).
I. High magnification of endothelial cells showing intraluminal filopodia (arrowheads) and Weibel-Palade bodies (arrows).
J. High magnification of an intermediate endothelial cell showing microtubules (arrowheads) and mural cell with its characteristic dilated RER (arrows) and abundant mitochondria (asterisk).
K. Mural cell (magenta) interdigitated with endothelial cells.
L. Composite images of ultrathin sections from the 17 GW cortex showing vascular distribution (magenta).
M-O Cortical blood vessels with dark (ED, green), intermediate (Ei, blue), and light (EL, purple) endothelial cells and a mural cell (magenta).
N. Higher magnification from M showing the junctional complexes between different types of endothelial cells (arrows) and the near obliterated vascular lumen (magenta line).
O. High magnification from N showing clathrin-coated vesicles (white arrowhead) and caveolae (black arrowhead) at the thin luminal surface.
P-Q. Blood vessels from the VZ at 17 GW (P) and 21 GW (Q) highlighting all 3 subtypes of endothelial cells and their mitochondria (arrows).
R-T. Higher magnification images to show the mitochondria and RER in each endothelial subtype (Light, R; Intermediate, S; Dark, T) at 17GW.
U. Quantification of mitochondrial surface area of each endothelial cell subtype at 17 and 21GW. Scale bars: A, L (5 μm); M, N: (2 μm); P, Q (1 μm); R-T (200 nm).
In U, each data point represents a blood vessel cell of the dark, intermediate, or light subtype from the 17 or 21 GW case. Statistics used ANOVA with post-hoc Student’s t-tests to assess differences within groups. Data are mean ± S.E.M.. Non-significant comparisons not shown.
On TEM, mural cells were closely associated with the endothelial cells, but not covering them in their entirety. These mural cells were characterized by electron-dense cytoplasm and irregular nuclei with deep invaginations, large nucleoli, and lumpy chromatin associated with the nuclear membrane (Figure 2B–C, J–K, Figure S2E, shaded in magenta). These mural cells contained abundant polyribosomes, RER and mitochondria (Figure 2C, J), and contacted endothelial cells using junctional complexes (Figure 2C), immature basement membranes (Figure 2G), and interdigitating cytoplasmic processes (Figure 2K, Figure S2E). Unlike the endothelial cells, the mural cells did not contain ultrastructural features that defined subtype specification (Table S1).
Profiling Second Trimester Brain Vascular Cells
To investigate the stages of vascular development during the second trimester, we used a FACS-based strategy to purify endothelial and mural cells. Based on single-cell transcriptomics data from prenatal human brain (Nowakowski et al., 2017), we identified CD31 (PECAM1), ANPEP (CD13), and CD45 (PTPRC) as an effective combination to isolate endothelial and mural cells from microdissected cortex and GE and simultaneously exclude perivascular myeloid cells (Figure 3A)(Crouch and Doetsch, 2018; Crouch et al., 2015). Flow cytometry confirmed the validity of these markers to isolate distinct populations of endothelial cells (CD45−;CD31+;ANPEP−) and mural cells (CD45−;CD31−;ANPEP+)(Figure 3A, Figure S3A). All CD31+ cells were CD34+, CD105lo, CD146+ and negative for PDGFR-β and ANPEP, whereas mural cells were PDGFR-β+, ANPEP+, CD146high, CD34−, and CD105lo (Figure S3B–C). To validate these results, we cultured endothelial and mural cells separately on collagen in endothelial growth media (EGM-2). In these conditions, endothelial cells displayed typical cobblestone morphology and expressed CD31 at cell-cell junctions, whereas mural cells had spindle-shape morphology and expressed PDGFR-β (Figure 3B). In 3D Matrigel cultures, both endothelial and mural cells formed tube-like structures as expected for vascular cells (Figure 3C–D). Next, we performed single-cell RNA sequencing (scRNA-seq) using FACS-isolated endothelial cells and mural cells from the 15–23 GW human GE and cortex (Figure 3E). In total, we collected data from 147,228 cells from 7 samples (2 regions each) with an average of 96,778 reads per cell, 1,468 genes per cell in sequencing depth, and 122,798 unique molecules per cell (Table S2). Visualization of scRNA-seq data from these samples with Uniform Manifold Approximation and Projection (UMAP) revealed distinct separation of endothelial and mural cells. Endothelial cells expressed PECAM1, CD34, and TIE1 and mural cells ANPEP, PDGFRB, and RGS5 as expected (Figure 3E–F). Contamination by other neural subtypes such as neuroblasts, astrocytes, microglia and oligodendrocytes was minimal (Figure S3D). While endothelial and mural cells from the two brain regions showed significant overlap on the UMAP, minor differences by age were noted, suggesting age-dependent developmental trajectories (Figure 3E).
Figure 3. Single-cell transcriptomics of endothelial and mural cells from prenatal human brain in the second trimester.
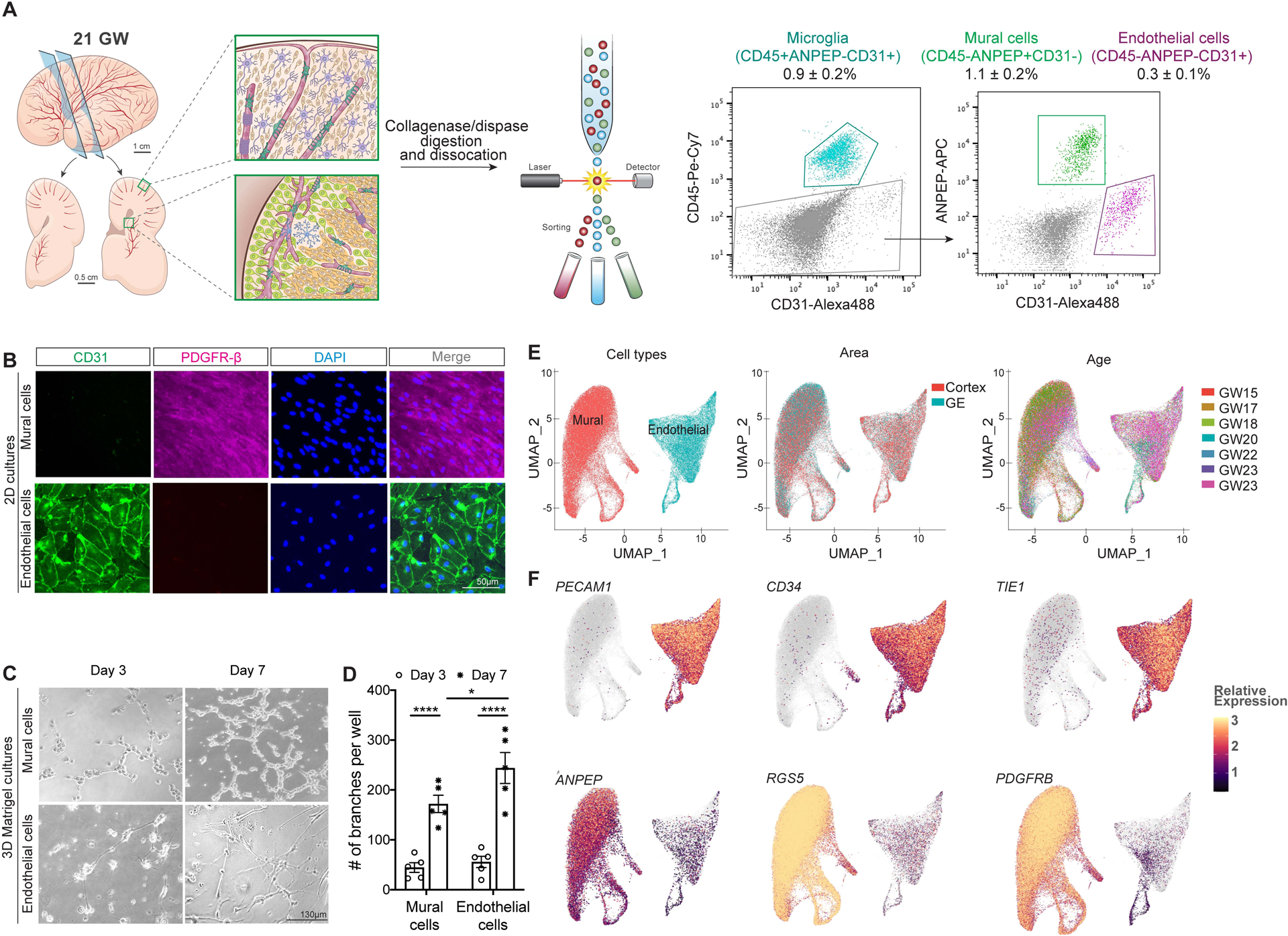
A schematic diagram for isolating endothelial and mural cells from the prenatal human brain. Left: Coronal sections of 21 GW prenatal human brain and higher power views depicting the cortex and GE and brief dissociation details. Right: Scattered dot plots showing the exclusion of CD45+ myeloid cells and selection of endothelial and mural cells.
B. Immunofluorescent images of CD31+ endothelial and PDGFR-β+ mural cells in 2D cultures in EGM-2 media.
C-D. Images of 3D Matrigel cultures of endothelial and mural cells showing their ability to form tubes (C). Quantification of the number of branch points established by endothelial and mural cells in 3D cultures (D). Each data point represents one well of cells from one case. Statistics used ANOVAs with post-hoc Student’s t-tests to assess differences within groups. Data are mean ± S.E.M. Non-significant comparisons not shown.
E. UMAP plots showing the clustering of endothelial and mural cells based on their subtypes, brain regions, and gestational ages.
F. Feature plots on the UMAP space showing the expression of pan-endothelial cell markers (PECAM1, CD34, and TIE1) and mural cell markers (ANPEP, RGS5, and PDGFRB) in each cluster.
Developmental Trajectories of Endothelial Cells
Unbiased clustering identified five subtypes of endothelial cells from 15–23 GW prenatal human brain, including mitotic, venous, capillary, tip cell, and arterial (Figure 4A–B, Table S3.) Genes enriched in the mitotic endothelial cell clusters included proliferative markers (TOP2A, CENPF, and BIRC5) and generated the gene ontology (GO) terms kinetochore, mitotic G2/M, AuroraB pathway, DNA replication, regulation of cell cycle process, and cell cycle (Figure S4A). In contrast, the arterial, venous, capillary, and tip cell clusters contained genes that were either shared with the P7 mouse brain or specific for prenatal human brain (Figure S4B)(Sabbagh et al., 2018). These included ALPL, CXCL12, ELN, GJA4, HEY1, SRGN, and UNC5B for the arterial endothelial cell cluster, ADM, ANGPT2, APLN, CXCR4, and PGF for the tip cell cluster, ADGRG6, IFI27, LY6E, PRCP, PMAIP1, RPS2 and RPS23 for the venous endothelial cell cluster, and CD27, CSRP2, HES1, PON2, SLCO1A2, and SLC39A10 for the capillary endothelial cell cluster.
Figure 4. Development of endothelial cells in the nascent vasculature of the second trimester human brain.

A. UMAP plots highlighting 15–23 GW endothelial cells according to subtypes and brain regions.
B. Heatmap showing mitotic, artery, tip cell, and capillary/venous gene expression in distinct groups.
C. Feature plots for representative genes for the mitotic, capillary/venous, tip cell, and arterial endothelial cells on the UMAP space.
D. RNA velocity analysis based on the scRNA-seq data of 15 and 23 GW endothelial cells indicating the stage-dependent lineage trajectories.
E-F. RNAscope for ADGRG6 (capillary/venous endothelial cell marker) in 15 and 23 GW human brain sections. Arrows indicate areas of the RNAscope signals.
G. Immunostaining for MFSD2A (capillary/venous endothelial cell marker) in 17 GW human brain sections. Arrow indicates vascular areas positive for the antibody staining.
H-I. RNAscope for ADM and ANGPT2 (tip cell markers) in 15 and 23 GW human brain sections. Arrows indicate vascular areas of the RNAscope signals. Note the absence of ADM and ANGPT2 in the cortical plate (Box 1), distant from the ventricular zone.
J. Immunostaining for ADM (tip cell marker, arrow) in 17 GW human brain sections.
K-L. RNAscope for GJA4 and FBLN5 (arterial markers) in 15 and 23 GW week human brain sections. Arrows indicate vascular areas of the RNAscope signals.
M. Immunostaining for ELN (arterial marker) in 17 GW human brain sections. Arrow indicates vascular areas positive for the antibody staining, and arrowhead indicates an ELN-negative area.
N. Volcano plot of differentially expressed genes in young (left side) versus old (right side) endothelial cells.
O. Gene ontology terms for the genes enriched in young (left) and old (right) endothelial cells.
P. FACS plots showing the selection of endothelial cells (left panel) and a histogram of Mitotracker Red expression in 16–19 GW, 21–23 GW, and adult cases (right panel).
Q. Quantification of low (103–104) and high (104–105) Mitotracker Red expression in endothelial cells from different gestational ages.
R-S. Quantification of OCR (oxygen consumption rate) and ECAR (extracellular acidification rate) in primary human brain endothelial cells. Values are normalized to starting point.
T. Quantification of the basal levels of OCR and ECAR in 16 and 24 GW human brain endothelial cells.
In Q, each data point represents a separate case. In R-S, points represent technical replicates of ≥ 4 wells of cells from the given gestational age. Statistics used ANOVA with post-hoc Student’s t-tests to assess differences within groups (Q) or Student’s t-tests (R-T). Data are mean ± S.E.M.. Non-significant comparisons not shown.
Next, we analyzed scRNA-seq data for endothelial cells using RNA velocity to decipher the relationships between different cell clusters and their developmental trajectories (La Manno et al., 2018). These results showed that at 15 GW mitotic endothelial cells gave rise to venous endothelial cells, which then progressed through an intermediate stage consisting of capillary endothelial cells and tip cells before terminating as arterial endothelial cells (Figure 4D, Figure S4C). At 23 GW, both venous and mitotic endothelial cells showed partial overlaps and were positioned at the beginning of the trajectory, giving rise to capillary endothelial cells, tip cells and arterial endothelial cells. The 23 GW samples showed more arterial and fewer capillary endothelial cells, supporting the maturation of vascular cells at this prenatal stage (Figure 4D).
Next, we performed RNAscope and immunostaining from the prenatal human brain at early (15–17 GW) and late (23–24 GW) second trimester sections. RNAscope showed that ADGRG6, a marker for venous and capillary clusters, was detected in non-continuous segments of CD31+ vasculature in pallial and subpallial VZ/SVZ at 15 and 23 GW (Figure 4E–F, left panel). In contrast, immunostaining showed diffuse presence of capillary/venous marker MFSD2A in endothelial cells (Figure 4G). Consistent with the results in Figure 1F–G, tip cell markers (ADM, ANGPT2) were focally enriched in endothelial cells in the pallial and subpallial VZ/SVZ, but sparsely present in endothelial cells in the cerebral cortex at 15 GW (Figure 4H). ADM and ANGPT2 expression persisted at 23 GW in endothelial cells in the VZ/SVZ, but was not detected in the endothelial cells in the cerebral cortex (Figure 4I). Consistent with the RNAscope data, immunostains for ADM confirmed its presence in endothelial cells in the VZ/SVZ (Figure 4J). Similar to the non-continuous expression of capillary/venous and tip cell markers, the arterial markers GJA4, FBLN5 and ELN were detected in discrete segments of the endothelial cells in larger blood vessels in the pallial and subpallial VZ/SVZ (Figure 4K–M).
Differential gene expression analysis showed that younger endothelial cells were more enriched in genes related to ribosomal functions and ribosomal subunits, and mitochondrial genes in oxidative phosphorylation, respiratory chain complex, cytochrome complex assembly, and antioxidant pathways (Figure 4N–O, Table S3). The over-representation of these metabolic pathways was consistent with the results from TEM, and supports that endothelial cells in 15–18 GW brain are metabolically different from adult endothelial cells, which use the glycolytic pathway as their primary energy source (Li et al., 2019). The metabolic pathways in the young endothelial cells resembled those in tumor endothelial cells (Rohlenova et al., 2020), suggesting that the younger endothelial cells might be exposed to a relatively hypoxic environment. In contrast, older endothelial cells showed enrichment of genes related to blood vessel development, chromatin-related genes, RNA processing, and structural genes, including cell junctions and supramolecular fiber organization (Figure S4C).
To characterize the mitochondrial contents and metabolism in the second trimester brain endothelial cells, we performed flow cytometry using Mitotracker Red and immunostains with Translocase of the outer mitochondrial membrane complex subunit 20 (TOMM20). These results showed that the endothelial cells from 15–18 GW had less mitochondrial mass than those from 20–23 GW (Figure S4D–E). Flow cytometry experiments with Mitotracker Red confirmed these results; endothelial cells from 15–18 GW had less abundant mitochondrial mass compared to adult endothelial cells, and trend toward less than those from 20–23 GW (Figure 4P–Q). In addition, Seahorse assays showed that, despite having lower basal levels of oxygen consumption rate (OCR) and extracellular acidification rate (ECAR), the endothelial cells from 15–18 GW exhibited more reserve capacity for respiration and less reserve capacity for glycolysis compared to those from 20–23 GW (Figure 4R–T). These results provide an important link between the morphology, transcriptomics, and functions of endothelial cells.
Mural cell development in the prenatal human brain
Unbiased clustering of our scRNA-seq dataset identified four subtypes of mural cells, including smooth muscle cells, classic pericytes, fibroblasts and mitotic mural cells (Figure 5A–B, Table S3). All four subtypes were detected in GE and cortex from 15 to 23 GW (Figure 5A). The mitotic mural cells expressed cell cycle-related genes, including AURKB, NUF2, BIRC5, KIF20B, UBE2C, and HHMR, which defined GO terms such as cell cycle, regulation of cell cycle process, and AuroraB pathway (Figure 5B–C, Figure S5A). Smooth muscle cells expressed genes such as ACTA2, TAGLN, MYL9, TPM1 and TPM2 (Figure 5B–C), with GO terms related to actin filament-based process, muscle structure development, smooth muscle contraction, and signaling by RhoGTPases (Figure S5A). In addition, our scRNA-seq data identified a subpopulation of mural cells which expressed marker genes for fibroblasts in adult mouse brain, such as COL1A1, COL3A1, LUM, and SERPING1 and as well as human-specific fibroblast marker genes like CLU, VCAN, and PTGDS (Figure S5B). The GO terms defined by these genes included fibroblast functions, extracellular matrix (ECM) organization, blood vessel development, and response to wounding (Figure 5B–C, Figure S5A). Finally, our scRNA-seq data identified classic pericytes, which expressed well-known pericyte genes KCNJ8, ATP1A2 and CD248 (Figure 5B–C). The GO terms for this classic pericyte subtype included ECM proteoglycans, basement membranes, and transport across the BBB, consistent with the known roles of these cells (Figure S5A)(Armulik et al., 2010; Cheng et al., 2018; Daneman et al., 2010).
Figure 5. Development of mural cells in the nascent vasculature of the second trimester human brain.

A. UMAP plots indicating 15–23 GW mural cells according to subtypes, brain regions, and prenatal ages.
B. Heatmap demonstrating mitotic, classic pericyte, smooth muscle cell, and fibroblast gene expression in distinct groups.
C. Feature plots showing representative genes for the mitotic, smooth muscle cell, classic pericyte, and fibroblast subtypes of mural cells on the UMAP space.
D. RNA velocity analysis based on scRNA-seq data of 15 and 23 GW mural cells indicating the stage-dependent lineage trajectories.
E-F. RNAscope for ATP1A2 and KCNJ8 (classic pericyte markers) in 15 and 23 GW human brain sections. Arrows indicate vascular areas where the RNAscope probe was localized.
G. Immunostaining for CD248 (classic pericyte marker) in 17 GW human brain sections. Note the gradient of expression highest at the ventricular surface.
H-I. RNAscope for LUM and SERPING1 (fibroblast markers) in 15 and 23 GW human brain sections. Arrows indicate vascular areas where the RNAscope probe was localized.
J. Immunostaining for LUM in 17 GW human brain sections.
K-L. RNAscope for MYL9 and TAGLN (smooth muscle cell markers) in 15 and 23 GW human brain sections. Arrows indicate vascular areas where the RNAscope probe was localized.
M. Immunostaining for MYL9 in 17 GW human brain sections.
Differential expression analysis showed that mural cells from 20–23 GW preferentially expressed genes associated with blood vessel development, cell adhesion, and ECM (Figure S5C–D, Table S4), consistent with their role as vascular support cells (Armulik et al., 2011; Attwell et al., 2016). In contrast, mural cells from 15–18 GW upregulated genes related to ribosomes, oxidative phosphorylation, smooth muscle cell differentiation, and immune-related groups like host interactions of HIV factors (Figure S5C–D). The upregulation of ribosomal genes in mural cells at 15–18 GW supported the more immature, progenitor state of mural cells at this age, whereas the presence of immune-related genes may relate to the emerging role of mural cells in the BBB (Daneman et al., 2010; Duan et al., 2018; Parker et al., 2020).
Next, we analyzed the trajectory of mural cells using RNA Velocity, which showed three distinct states of cellular maturation in mural cells at 15 GW, beginning with mitotic mural cells (brown, UBE2C+, BIRC5+), which progressed towards non-proliferative smooth muscle cells (red, MYL9+, TAGLN+), and finally to classic pericytes (blue, ATP1A2+, KCNJ8+)(Figure 5D, for feature plots see Figure S5E). In addition, 15 GW mural cells also contained a large group of fibroblasts that expressed LUM (yellow) and SERPING1 (Figure 5D, Figure S5E). Although a subset of fibroblasts seemed to arise from mitotic mural cells, the majority of fibroblasts at this age appeared to derive from cells of unknown origin. Consistent with the progressive reduction in Ki-67+ mural cells from 14–17 GW to 21–25 GW (Figure 1K), RNA velocity analysis in the 23 GW mural cells showed a marked reduction of the mitotic mural cell cluster, a distinct population of smooth muscle cells that gave rise to fibroblasts, and expansion of classic pericytes (Figure 5D right panel and Figure S5E).
Finally, RNAscope detected the expression of classic pericyte markers KCNJ8 and ATP1A2 in many mural cells in smaller blood vessels in the VZ/SVZ of pallium and subpallium at 15 and 23 GW (Figure 5E–F, Movie S3). While KCNJ8 expression was restricted to mural cells, ATP1A2 transcripts were detected in neural progenitors in the ventricular zone (Figure 5E)(Fu et al., 2021; Pollen et al., 2015; Smith et al., 2021). Immunostains for another classic pericyte marker CD248 showed CD248+ cells encasing endothelial cells in the subpallial VZ/SVZ at 17 GW (Figure 5G). These results were consistent with those from NG2 (Figure 1E), another classic pericyte marker, and suggested a gradient of classic pericytes developing from the ventricular wall. Unlike pericytes, RNAscope for fibroblast markers LUM and SERPING1 showed that these fibroblasts were more readily detected around the blood vessels of the cerebral cortex in 15 and 23 GW, and sparsely present in the vasculature in the pallial and subpallial VZ/SVZ (Figure 5H–I). As previously noted (Eze et al., 2021), abundant LUM transcripts were found in neural progenitors in the VZ/SVZ at 15 GW. Immunostains also showed that LUM proteins were broadly present in the ventricular zone and on rare perivascular cells (Figure 5J). Finally, RNAscope for smooth muscle cell markers, MYL9 and TAGLN, showed an infrequent presence of these cells at 15 and 23 GW. At 15 GW, MYL9+ and TAGLN+ cells were most commonly found in larger vessels of the cortex (Figure 5K–L, panels 1). Immunostains detected few MYL9+ smooth muscle cells around the vasculature in the pallial and subpallial VZ/SVZ (Figure 5M).
Molecular mechanisms supporting vascular development
Next, we used CellChat to query the signaling pathways for cell-cell communication between endothelial and mural cells (Jin et al., 2021). The most over-represented pathways were ECM protein families, such as collagen, laminin, and fibronectin (FN)(Figure 6A, Figure S6A–B). Chord plots and violin plots showed extensive expression of Collagen type IV α1 chain (COL4A1) and Integrin α1 (ITGA1) in most vascular subtypes at 15 and 23 GW (Figure 6B). In agreement, immunostains revealed high levels of COL4A1 and ITGA1 throughout the brain vasculature at 23 GW (Figure 6C). Unlike COL4A1, COL1A1 was only expressed by fibroblasts at 15 and 23 GW, whereas COL9A3 showed expression in tip cells only (Figure S6A). Other adhesion or tight junction proteins included JAM (Junction Adhesion Molecules), Cadherins (CDH), PECAM1, Occludin (OCLN), and Claudin (CLDN), though their contribution was small (Figure 6A, Figure S6C). In addition, CellChat identified ligand-receptor pairs, such as Notch, vascular endothelial growth factor (VEGF), PDGF, Ephrins, and insulin-like growth factor, known to promote vascular expansion and remodeling (Figure 6A and Figure S6D–F). However, the most abundant growth factor-related pathway was Midkine (MDK), which promotes angiogenesis and is structurally and functionally similar to pleiotrophin (Ross-Munro et al., 2020; Weckbach et al., 2012)(Figure 6D). To investigate its expression in the developing human brain, we performed immunostains for MDK and two of its receptors, Syndecan 2 (SDC2) and Integrin β1 (ITGB1). Consistent with the scRNA-seq data, ITGB1 showed extensive expression in all types of endothelial and mural cells (Figure 6E and Figure S6E). In contrast, SDC2 expression showed progressive increase in smooth muscle cells from 15 to 23 GW and in mitotic mural cells and fibroblasts at 23 GW (Figure 6D, F and Figure S6E). Finally, 3D Matrigel tube assays showed that MDK (20 ng/ml) increased, whereas MDK inhibitor iMDK (100 nM) reduced, branching in both endothelial and mural cells (Figure 6G–I).
Figure 6. Molecular pathways for cell-cell communication between endothelial and mural cells in the prenatal human brain.

A. Rank order of significant ligand-receptor pairs utilized by the endothelial and mural cells in the cell-cell communication in the second trimester human brain.
B. (Top panels) Chord plots summarizing expression of collagen genes and their receptors by endothelial and mural cells at 15 and 23 GW. (Bottom panels) Violin plots of COL4A1 and ITGA1 in vascular subtypes.
C. Confocal images showing immunostains for COL4A1, ITGA1, and CD31 in the ventricular zone of 23 GW prenatal human brain.
D. (Top panels) Chord plots summarizing expression of midkine and its receptors by endothelial and mural cells at 15 and 23 GW. (Bottom panels) Violin plots of MDK, SDC2, and ITGB1 in vascular subtypes.
E. Confocal images showing immunostains for MDK, ITGB1, and CD31 in the ventricular zone of 23 GW prenatal human brain.
F. Confocal images showing immunostains for MDK, SDC2, and CD31 in the ventricular zone of 23 GW prenatal human brain.
G. 3D Matrigel cultures of endothelial and mural cells in the presence of MDK or iMDK. Images shown at 3 and 6 hours.
H-I. Quantifcation of number of branches in Matrigel cultures at 3, 6, and 12 hours in endothelial or mural cells with MDK or iMDK. Each data point represents the average of 3 wells from a separate biological replicate. Statistics used ANOVA with post-hoc Student’s t-tests to assess differences within groups. Upper asterisk compares between control and MDK; lower asterisk between control and iMDK. Data are mean ± S.E.M.. Non-significant comparisons not shown.
Angiogenesis in Cerebral Organoids
To examine the roles of vascular cells in neural progenitors, we transplanted FACS-purified endothelial and mural cells into 7–9 week-old human induced pluripotent stem cell (iPSC)-derived cortical organoids, which mimic cortical development during the second trimester (Bhaduri et al., 2020). Prior to transplantation, endothelial and mural cells were labeled by AAV-CMV-GFP to track their presence in the organoids (Figure 7A–B). Two weeks after transplantation, immunostains showed that 27% of the transplanted endothelial cells and 22% of mural cells were Ki-67+ (Figure 7C–D), and RNAscope identified different mural and endothelial cell subtypes, including smooth muscle cells (MYL9 and TAGLN), classic pericytes (ATP1A2 and KCNJ8), fibroblasts (SERPING1 and LUM), tip cells (ANGPT2 and ADM), arterial (GJA4 and FBLN5), and capillary/veinous (ADGRG6 and PRDX3). These results showed that the majority of the mural cells in the cerebral organoids displayed smooth muscle cell subtype markers with smaller percentages of classic pericytes and fibroblasts (Figure 7E–F). In contrast, most transplanted endothelial cells expressed tip cell markers, ANGPT2 and ADM (Figure 7G–H).
Figure 7. Human endothelial and mural cells promote neurogenesis and reduce cell stress in cortical organoids.
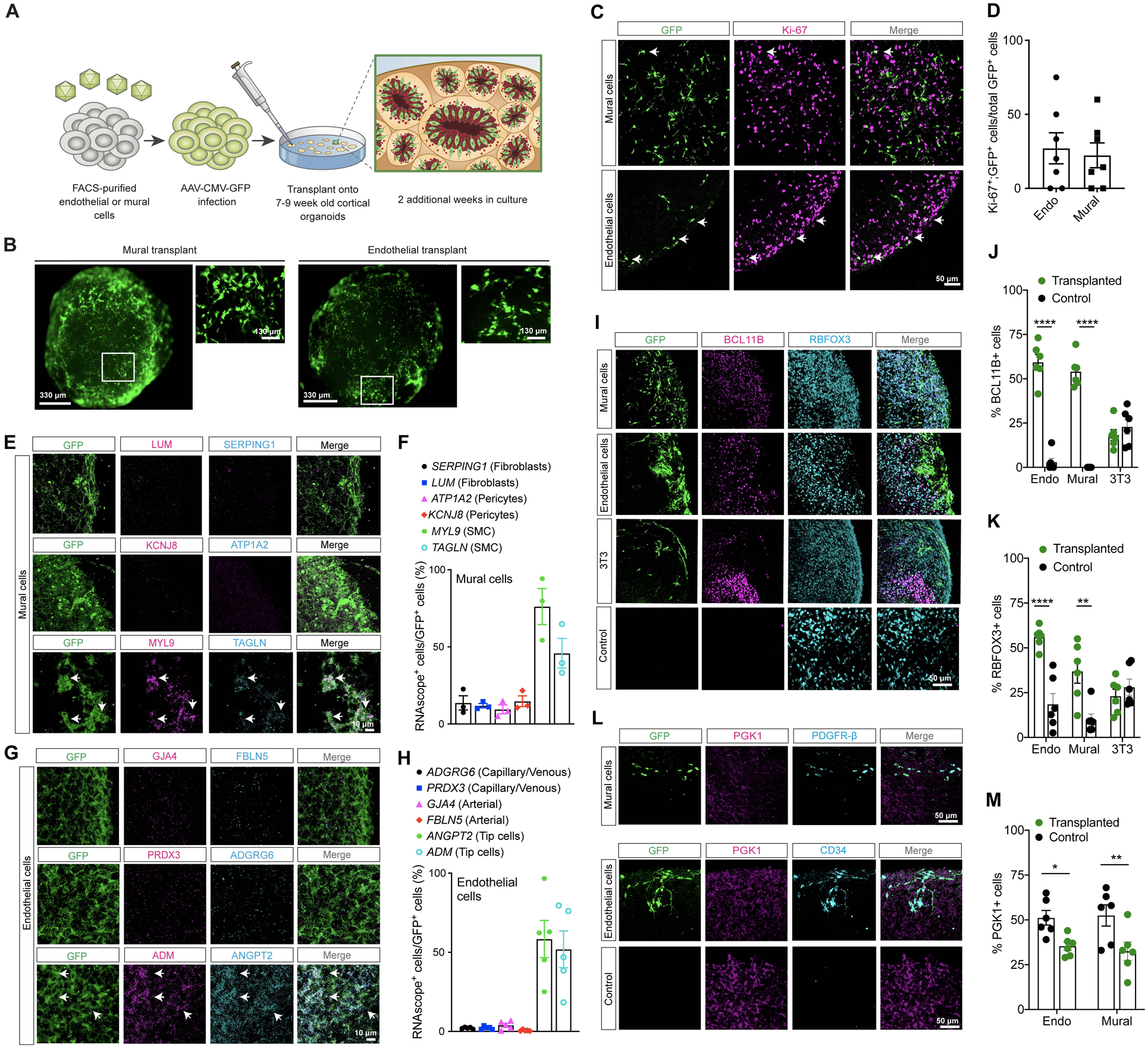
A. Schematic diagram for transplanting AAV-GFP labeled endothelial or mural cells into cortical organoids.
B. Microscopic images of transplanted endothelial and mural cell organoids after 7 days in culture.
C. Confocal images for immunostains for GFP and Ki-67 in the transplanted organoids.
D. Quantification of Ki-67+ cells in transplanted organoids.
E. Confocal images showing LUM, SERPING1, KCNJ8, ATP1A2, MYL9 and TAGLN RNA in transplanted mural cells.
F. Graph showing the subtype of transplanted mural cells in the cortical organoids as determined by RNAscope probes.
G. Confocal images of PRDX3, ADGRG6, GJA4, FBLN5, ANGPT2, and ADM RNA in transplanted endothelial cells.
H. Graph showing the subtype of transplanted endothelial cells in the cortical organoids as determined by RNAscope probes.
I. Confocal images for GFP, BCL11B (CTIP2), and RBFOX3 (NeuN) in the transplanted organoids.
J-K. Quantification of RBFOX3+ or BCL11B+ cells in organoids in GFP+ or control regions.
L. Confocal images for GFP, PGK1, and CD34 (endothelial cells) or PDGFR-β (mural cells) in organoids.
M. Quantification of PGK1+ cells in the organoids in GFP+ or control regions.
In D, J, K, and M, each data point represents the average of 5 sections from one transplant. Statistics used Student’s t-tests. Data are mean ± S.E.M.. Non-significant comparisons not shown.
To determine how transplanted vascular cells affect neuronal development, we quantified the number of neurons expressing BCL11B (CTIP2) (which labels excitatory neurons in layers 5–6 of the cortex), CUX1/2 (excitatory neurons in layers 2–4 of the cortex), SATB2 (callosal projection neurons), RBFOX3 (NeuN), and PAX6 (progenitors) within 350 μm of GFP+ endothelial cells or mural cells (Figure 7I–K, Figure S7A–E). As controls, we quantified areas without transplanted cells. In addition, we transplanted NIH3T3 cells into the organoids as cell type-specific controls. These results showed that transplanted mural and endothelial cells increased the number of NeuN+ cells and BCL11B+ cells in the organoids (Figure 7I–K). Compared to endothelial cells and NIH3T3 cells, the transplanted mural cells modestly promoted the development of SATB2+ neurons, whereas neither endothelial cells, mural cells or NIH3T3 cells affected the development of CUX1/2+ or PAX6+ cells (Figure S7A–E).
Our previous work shows that newly born neurons in cerebral organoids exhibit cellular stress and upregulate genes in the endoplasmic reticulum (ER) stress pathway, including PGK1, ARCN1 and GORASP2. Immunostains for PGK1 showed that both endothelial and mural cells decreased the number of PGK1+ cells in regions adjacent to the vascular cells (Figure 7L–M). In addition, immunostains for glycolytic enzymes enolase and GORASP1, which are readouts for caspase-mediated Golgi fragmentation (Gee et al., 2011; Kim et al., 2016), showed that transplanted endothelial and mural cells can reduce the number of enolase+ and GORASP1+ cells in the organoids (Figure S7F–I).
Discussion
One remarkable discovery of our study is the mosaic development of vascular cells in the prenatal human brain (Figure S7J). Specifically, our results support a model wherein the developing vasculature in the second trimester is tiled by an ensemble of diverse endothelial and mural cell subtypes, including those previously reported in postnatal mouse brain (Sabbagh et al., 2018; Vanlandewijck et al., 2018). Consistent with this model, RNA velocity of endothelial cell transcriptomes shows a directional flow from immature venous and mitotic endothelial cells, through capillary and tip cells, ending with arterial endothelial cells (Figure 4). In agreement with this trajectory, endothelial cells expressing arterial markers GJA4 and FBLN5 were present in low abundance in the prenatal human brain at 15 GW, and became more prominent at 23 GW. Conversely, both tip cells and capillary endothelial cells show progressive reductions in their relative abundance from 15 to 23 GW. In addition to these temporal changes, endothelial cell development may also be influenced by environmental cues. For example, the ventricular zone is largely occupied by tip cells, mitotic endothelial cells, and mitotic mural cells. In this region, endothelial tip cells have previously been shown to promote neurogenesis using a VEGF-mediated mechanism (Di Marco et al., 2020). Transplanted FACS-isolated endothelial cells also preferentially displayed a tip cell phenotype under the neurogenic environment of cerebral organoids. Together, these results support the idea that the developmental trajectories of endothelial cells are guided by an intrinsic program and can be influenced by environmental signals that encourage a specific subtype.
Another intriguing result revealed by our scRNA-seq datasets is that the endothelial cells from early second trimester exhibit more abundant transcripts related to oxidative phosphorylation. Using TEM, confocal microscopy and FACS, we show that the endothelial cells from 17 GW have less mitochondrial mass compared to those from 21–22 GW and adult brain (Figures 2P–U, 4P–Q, S4D–E). In addition, endothelial cells from 15–18 GW exhibit more reserve capacity for respiration and less reserve capacity for glycolysis compared to those from 20–23 GW (Figure 4R–T). These results support the idea that the endothelial cells from early second trimester may upregulate the expression of genes related to oxidative phosphorylation to compensate for smaller mitochondrial masses. Future studies using subtype-specific markers will shed light on how these ultrastructural and bioenergetic properties of endothelial cells correlate with specific transcriptomic subpopulation(s).
Three distinct subtypes of mural cells, including classic pericytes, fibroblasts, and smooth muscle cells, have been identified in the adult mouse brain. Our approach identifies these three mural cell subtypes and an additional subpopulation of mitotic mural cells in the prenatal human brain. To query the developmental trajectories of mural cells, RNA velocity at 15 GW shows the mitotic mural cells can give rise to fibroblasts and smooth muscle cells. At this age, classic pericytes represent a very small population that are derived from smooth muscle cells. Furthermore, this analysis suggests that a subpopulation of fibroblasts may also function as progenitors. By 23 GW, the relative abundance of mitotic mural cells reduces significantly. Instead, the smooth muscle cells are projected to give rise to classic pericytes and fibroblasts, both representing the more abundant cell types at this age.
Contrary to the lack of smooth muscle cells in embryonic mouse brain (Jung et al., 2018), our results reveal cells expressing smooth muscle cell markers in the second trimester human brain vasculature. These results highlight species-specific differences in the timing of smooth muscle cell development in prenatal human vasculature. Notably, our mural cell lineage trajectory differs from recent reports in the developing mouse heart and postnatal mouse brain (Ando et al., 2022; Volz et al., 2015). Future studies using lineage tracing and functional characterization will be critical to understand organ and species-specific differences in mural cell fate.
Our CellChat analysis provides important insights into the molecular mechanisms by which these endothelial and mural cells interact to promote prenatal human brain angiogenesis. Among the most prominent ligand-receptor pairs are collagen and laminin, which are related to ECM organization. These results are supported by immunostaining and TEM which confirm the presence of vascular basal laminae. Interestingly, the thin and discontinuous basement membrane in TEM suggests that the second trimester is an early and dynamic period of BBB formation. Another top ligand-receptor pair for endothelial-mural cell interactions is MDK and its receptors, integrins and syndecan. MDK has also been investigated as a key mechanism that promotes angiogenesis induced by hypoxia and in tumorigenesis (Filippou et al., 2020; Weckbach et al., 2012). In support of these findings, our results comparing the transcriptomes of endothelial cells at 15–18 GW and 20–23 GW show that the metabolic pathways employed by these young endothelial cells resemble those in tumor endothelial cells (Rohlenova et al., 2020). Together, these findings capture an important metabolic mechanism employed by endothelial cells in response to the relatively hypoxic environment in the prenatal human brain.
Our results in Figure 1 show that angiogenesis is actively occurring in the VZ and SVZ, where robust neurogenesis exists. We previously showed that organoids capture many aspects of neurogenesis in the ventricular zone of the prenatal human cerebral cortex from 6–22 GW (Bhaduri et al., 2020). Consistent with these findings, these organoids promote the differentiation of transplanted endothelial and mural cells toward tip cells and smooth muscle cells, respectively. While unexpected, these results reveal how a highly neurogenic environment, such as the cortical organoids, can impact the differentiation of vascular cells. While the exact mechanism(s) remain unclear, our results suggest that the VEGF-VEGFR signaling pathway and actin filament-based processes may promote the development of tip cells and smooth muscle cells, respectively. Another important finding of our study is that the transplanted vascular cells can improve neurogenesis and reduce cellular stress in the cortical organoids (Figure 7 and Figure S7). One potential mechanism for the transplanted vascular cells to reduce cellular stress is endothelial nitric oxide synthase (eNOS), which has been shown to protect against brain injury after ischemia or TNFα-induced cytotoxicity (De Palma et al., 2008; Faraci, 2006). Since vascularization of brain organoids is an increasingly popular area of research, our findings will open new directions to improve the environmental milieu for cortical organoids and to reveal pathways that govern angiogenesis and neurogenesis in the prenatal human brain.
Finally, congenital vascular diseases have profound impacts on neurodevelopment. For example, mutations in COL4A1 are associated with significant neonatal hemorrhages and subsequent leukoencephalopathy (Gould et al., 2005). Indeed, collagen and laminin are the two most dominant signaling pathways in second trimester human brain vascular cells, and COL4A1 specifically is expressed in all of our vascular subtypes. These results provide important insights as to why vascular defects caused by mutations in COL4A1 arise as early as the second trimester. In contrast, genes involved in the formation of adherens and tight junctions only show limited expression at this age. Our data, available at https://vascular-dev.cells.ucsc.edu, will be a resource for future studies of vascular cell subtypes and therapeutic strategies to facilitate the maturation of the brain vasculature in premature infants.
Limitations of this study
Our results provide an important blueprint for additional studies that focus on vascular development in the third trimester, perinatal and early postnatal stages. These results will progress to a more complete model of vascular cell changes and environmental influences in other critical periods. However, there are several limitations to our study. First, despite the differences in the vascular density between the VZ and SVZ in the pallium and subpallium during the second trimester, our single-cell transcriptomic data did not show any definitive differences in endothelial or mural cells from these regions in morphology, transcriptomes, or subtype classifications. It is possible that region-specific factors from non-vascular cells may dictate the timing and the extent of angiogenesis in different parts of the developing brain. Another limitation of our study is that organoids appear to restrict subtype development of the transplanted vascular cells. Future organoid experiments could benefit from including additional cellular repertoires to more faithfully model prenatal human brain development. Finally, recent studies show that BBB can be targeted by neuroinflammatory or neurodegenerative conditions, but how BBB is established in normal human brain remains unclear. Future studies on BBB development in the third trimester and postnatal stages will provide important connections with our findings in the prenatal human brain.
STAR Methods
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact Eric.huang2@ucsf.edu
Materials availability
Reagents generated in this study are available, upon reasonable requests, with a completed material transfer agreement.
Data and code availability
Single-cell RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Any FACS or microscopy data, confocal, electron, or other, reported in this paper will be shared by the lead contact upon request.
No original code was generated in this manuscript. All previously published algorithms are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key resources table
| REAGENT or RESOURCE | SOURCE | RRID IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-human CD131 | Agilent | AB_2892053 |
| Rabbit anti-human NG2 | Millipore | AB_11213678 |
| Goat anti-human PDGFR-β | R&D | AB_355339 |
| Rabbit anti-human Ki-67 | BD Biosciences | AB_396287 |
| Mouse anti-human CD31-Alexa488 | BD Biosciences | AB_647081 |
| Mouse anti-human CD13-APC | BD Biosciences | AB_398624 |
| Mouse anti-human CD45-PeCy7 | BD Biosciences | AB_396854 |
| Mouse anti-human CX3CR1-Alexa647 | BD Biosciences | AB_2869728 |
| Mouse anti-human PDGFR-β-PE | BD Biosciences | AB_397132 |
| Mouse anti-human CD105-PE | BD Biosciences | AB_2033932 |
| Mouse anti-human CD34-PE | BD Biosciences | AB_10563214 |
| IgG2a, K Mouse, Alexa Fluor 488, Clone: eBM2a, Isotype Control | Thermo Fisher | AB_470233 |
| APC Mouse IgG1, k Isotype Control | BD Biosciences | AB_398613 |
| PE Mouse IgG1, k Isotype Control | BD Biosciences | AB_396091 |
| PE-Cy7 Mouse IgG1 k Isotype Control | BD Biosciences | AB_396914 |
| Rabbit anti-human MFSD2A | ThermoFisher | AB_11152332 |
| Rabbit anti-human LUM | ThermoFisher | MA5–29402 |
| Rabbit anti-human GJA4 | Abcam | AB_2861171 |
| Mouse anti-human ADM | Thermo Fisher | AB_2722821 |
| Mouse anti-human smooth muscle actin | Sigma | AB_476701 |
| Goat anti-human COL4A1 | Millipore | AB_92262 |
| Rabbit anti-human SDC2 | Sigma | AB_2678934 |
| Rabbit anti-human ITGB1 | Sigma | AB_2732634 |
| Chicken anti-human GFP | Abcam | AB_300798 |
| Guinea Pig anti-human RBFOX3 (NeuN) | Fisher | AB_11205592 |
| Rat anti-human BCL11B (CTIP2) | Abcam | AB_2064130 |
| Rabbit anti-human PGK1 | Thermo Fisher | AB_2161216 |
| Sheep anti-human CD34 | R&D | AB_10973459 |
| Goat anti-human Midkine | R&D | AB_2143400 |
| Rabbit anti-human ITGA1 | Abcam | Catalog number ab181434 |
| TOMM20 | Abcam | AB_2889972 |
| CUX1/2 | Abcam | AB_941209 |
| SATB2 | Abcam | AB_882455 |
| PAX6 | Abcam | AB_2750924 |
| Enolase | Abcam | AB_2827927 |
| GORASP2 | Protein Tech | AB_2113473 |
| Bacterial and virus strains | ||
| AAV-CMV-GFP | Vector Labs | Catalog number 7117 |
| Chemicals, peptides, and recombinant proteins | ||
| Triton-X | Sigma | Catalog number T9284 |
| Tris-HCL | Corning | Catalog number 46-031-CM |
| Blocking reagent | Perkin Elmer | Catalog number FP1012 |
| BSA | Sigma | Catalog number A7979-50ML |
| Glucose | Sigma | Catalog number G7021-1KG |
| DNase | Worthington | Catalog number LS002139 |
| Collagenase/dispase | Sigma | Catalog number 11097113001 |
| Percoll | Signa | Catalog number P1644-500ML |
| Dapi | Thermo Fisher | Catalog number D1306 |
| Growth-Factor reduced Matrigel | Corning | Catalog number 356230 |
| Type 1 collagen | Stem Cell Technologies | Catalog number 4902 |
| Rock Inhibitor | Stem Cell Technologies | Catalog number 72304 |
| IWR1-endo | cayman chemical | Catalog number 13659 |
| SB431542 | Tocris | Catalog number 1614 |
| Mitotracker Red | ThermoFisher | Catalog number M7512 |
| Recominant Midkine | EMD Millipore | Catalog number AG243 |
| Midkine inhibitor | Axon Medchem | Catalog number 2258 |
| Critical commercial assays | ||
| RNAscope Multiplex Fluorescence V2 Assay | ACD | Catalog number 323135 and 323180 |
| Chromium Single Cell 3’ GEM, Library and Gel Bead Kit V3 | 10x Genomics | Catalog number 1000075 |
| Chromium Chip B Single Cell Kit, 48 reactions | 10x Genomics | Catalog number 1000153/1000073 |
| Deposited data | ||
| Raw and analyzed prenatal human vascular data | This paper | GEO: GSE63473 |
| Experimental models: Cell lines | ||
| Human umbilical vein endothelial cells | ATCC | Catalog number CRL-1730 |
| NIH 3T3 | ATCC | CVCL 0594 |
| 1323-4 iPSC line | Gladstone Institute | Bruce Conklin lab |
| H28126 iPSC line | University of Chicago | Gilad |
| Oligonucleotides | ||
| RNAscope® Probe – Hs-ATP1A2-C2 | ACD | Catalog number 497451-C2 |
| RNAscope® Probe – Hs-TAGLN-C2 | ACD | Catalog number 498961-C2 |
| RNAscope® Probe – Hs-KCNJ8 | ACD | Catalog number 521891 |
| RNAscope® Probe – Hs-SERPING1 | ACD | Catalog number 869491 |
| RNAscope® Probe – Hs-PRDX3 | ACD | Catalog number 1062541-C1 |
| RNAscope® Probe – Hs-LUM-C2 | ACD | Catalog number 494761-C2 |
| RNAscope® Probe – Hs-ANGPT2-C2 | ACD | Catalog number 594511-C2 |
| RNAscope® Probe – Hs-FBLN5-C2 | ACD | Catalog number 593511-C2 |
| RNAscope® Probe – Hs-ADGRG6-C2 | ACD | Catalog number 480121-C2 |
| RNAscope® Probe – Hs-MYL9 | ACD | Catalog number 593741 |
| RNAscope® Probe – Hs-GJA4 | ACD | Catalog number 856221 |
| RNAscope® Probe – Hs-ANXA2 | ACD | Catalog number 855281 |
| RNAscope® Probe – Hs-ADM-C1 | ACD | Catalog number 582511 |
| Software and algorithms | ||
| Fiji | Schneider et al., 2012 | RRID:SCR_002285 |
| Imaris | Oxford Instruments | RRID:SCR_007370 |
| Prism | GraphPad Prism Version 8.4.3 | RRID:SCR_002798 |
| FlowJo | FlowJo Version 10.6.2 | RRID: SCR_008520 |
| RNAVelocity | Velocyto.py Version 0.17 and scvelo Version 0.2.4 | RRID:SCR_018167 |
| Adobe Illustrator | Adobe | RRID:SCR_010279 |
| R | The R foundation | RRID:SCR_001905 |
| RStudio | The R foundation | RRID:SCR_000432 |
| Cell Chat | CellChat Version 1.1.3 | RRID:SCR_021946 |
| Other | ||
| Cell browser for cells in the developing human brain | This paper | https://cells.ucsc.edu/?ds=vascular-dev |
Experimental Model and Subject Details
Human tissue samples
De-identified tissue samples were collected with previous patient consent in strict observance of the legal and institutional ethical regulations. Autopsy consent and all protocols were approved by the Human Gamete, Embryo, and Stem Cell Research Committee (institutional review board) at the University of California, San Francisco. All cases were determined by chromosomal analysis, physical examination, and/or pathological analysis to be control tissues, which indicates that they were absent of neurological disease. Cases with any abnormalities in these parameters were not used for this manuscript. The same cases were able to be used for immunohistochemistry and RNAscope. For TEM, scRNA-seq, Matrigel tube assays, and organoid transplants, none of the same cases were used for two types of experiments because the tissue or cells needed to be processed in a different way for each type of experiment.
| Experiment | Cases |
|---|---|
| Immunohistochemistry and RNAscope validations | GW 14, 15, 17, 18, 21, 21, 22, 23, 38, 39, 39, 39 |
| TEM | GW 17, 21, 23 |
| scRNA-seq | GW 15, 17, 18, 20, 22, 23, 23 |
| Matrigel tube assay | GW 17, 21, 21, 22, 23 |
| Mitotracker Red experiments | GW 16, 17, 18, 19, 21, 21, 22, 23. Adult cases: 62Y, 62Y, 65Y. |
| Seahorse assay | GW 16, 24 |
| Organoid transplants | GW 13, 18, 18, 20, 21, 23 |
Method Details
Human tissue collection.
As previously described (Nowakowski et al., 2017; Paredes et al., 2016, 2022), human specimens with post-mortem interval of less than 48 hours were collected with previous patient consent in strict observance of the legal and institutional ethical regulations. Protocols were approved by the Human Gamete, Embryo, and Stem Cell Research (GESCR) Committee at the University of California, San Francisco. Tissue samples were dissected in artificial cerebrospinal fluid containing 125 mM NaCl, 2.5 mM KCl, 1mM MgCl2, 1 mM CaCl2, and 1.25 mM NaH2PO4 under a stereotaxic dissection microscope (Leica).
Immunohistochemistry.
Brains were cut into ~1.5 cm coronal or sagittal blocks, fixed in 4% paraformaldehyde for 2 days, and then cryoprotected in a 30% sucrose solution. Blocks were cut into 30-micron sections on a cryostat, mounted on glass slides for immunohistochemistry, and stored at −80C. Frozen slides were gradually equilibrated to room temperature, moved from −80°C to 4°C the night prior to staining and then to the lab bench for 2 hours before beginning the immunostaining protocol. Slides were washed once with 1X PBS for 5 minutes, then once with 1X TBS for 5 minutes before blocking with TBS++++ (goat serum, BSA, albumin, glycine and triton X in TBS) for 1 hour. Primary antibody incubations were performed overnight at room temperature in TBS++++. The following day, three 1x TBS washes were performed before incubating with secondary antibodies in TBS++++ for 2 hours. After three additional TBS washes, DAPI was added and the slides were mounted. Ki-67 staining required antigen retrieval and TSA amplification as previously described (Paredes et al., 2016). The autofluorescence caused by red blood cells in postmortem brain tissues generated significant with the RNAscope workflow and were removed from the images with post-image processing in ImageJ. Raw images available upon request.
Vascular quantifications.
To quantify the surface area of vascular cells in the developing human brain, images were acquired using a Keyence VH-600 digital microscope with mouse anti-human CD31, goat anti-human PDGFR-β, and rabbit anti-human NG2. The entire ventricular area was captured. Zones were determined based on cellular features in the ganglionic eminences (GE). Zone 1 is the ventricular zone with dense radial glia fibers, and Zone 2 includes densely packed newly-born neurons, which are PDGFR-β negative. Areas lateral to Zone 2 are designated as Zone 3. For consistency, the same measurements were used for the GE and the cortex in each slide. To determine the surface area occupied by the vasculature, the particle analysis tool in ImageJ was used. Filopodia and number of branches in each zone were counted manually from 40X confocal images of each zone from the same slides. To determine mitochondrial surface area, sections were stained with CD34 and Translocase of the outer mitochondrial membrane complex subunit 20 (TOMM20). In ImageJ, the non-vascular areas were subtracted from the image and we measured the area of all mitochondria and divided by the surface area of the CD34+ staining in that section.
Transmission Electron Microscopy.
For transmission electron microscopy (TEM), samples were sectioned with a vibratome (200 microns), postfixed with 2% osmium tetroxide solution. Sections were dehydrated in crescent ethanol concentrations, stained with 2% uranyl acetate, and finally embedded in araldite resin (Durcupan ACM Fluka, Sigma), and polymerized at 69 °C for 72 hours. Ultrathin sections were obtained (70 nm) and were contrasted with lead citrate solution on the grids. Postfixation was performed with 7% glucose-1% Osmium tetroxide and, afterwards, followed a conventional embedding protocol. Vascular cells were identified based on their subcellular features and location. Similar to confocal microscopy, mitochondrial surface area was determined by measuring the area of each mitochondria and dividing by the surface area of the endothelial cell in that section. For quantification in Figure 2U, 30 blood vessel segments were quantified per each subtype per case.
Fluorescence-Activated Cell Sorting.
To isolate vascular cells from prenatal human brain tissues, we adapted a previously published protocol that was used for the adult mouse brain (Crouch and Doetsch, 2018; Crouch et al., 2015). Briefly, brain tissue was minced with a scalpel and digested with collagenase/dispase (3 mg/ml) for 30 min at 37°C with rotation, triturated in 2% FBS in 1X PBS with DNase (0.25 mg/ml), and centrifuged through 22% Percoll to remove debris. The samples were incubated on ice for 15 min with Alexa488-conjugated mouse anti-human CD31 to label endothelial cells (1:100), Allophycocyanin (APC)-conjugated mouse anti-ANPEP (1:100) to label mural cells, and PECy7-conjugated mouse anti-hCD45 (1:200) to exclude blood cells and microglia. After washing, samples were resuspended in HBSS buffer with DAPI (1:500) to exclude dead cells. Cells were sorted using a Becton Dickinson FACSAria using 13 psi pressure and 100 μm nozzle aperture. All FACS gates were set using unlabeled cells, single- color and isotype controls from human samples. For flow cytometry experiments to examine other vascular markers, the same procedure was used with the following antibody combinations: Phycoerythrin (PE)-conjugated mouse anti-hPDGFR-β (1:200), Phycoerythrin (PE)-conjugated mouse anti-hCD146 (1:200), Phycoerythrin (PE)-conjugated mouse anti-hCD34 (1:200), and Phycoerythrin (PE)-conjugated mouse anti-hCD105 (1:100). Mitotracker Red was added at 50 nM.
Vascular cell cultures.
Endothelial or mural cells from cortex were pooled from 4 – 6 FACS experiments and cultured in EGM-2 media (Lonza) on collagen type 1-coated wells in 24 well dishes for 2D experiments. 3D experiments were performed in Ibidi μ-Slide Angiogenesis with 20,000 cells per well in undiluted Growth-Factor reduced Matrigel (Corning) with EGM-2 media for 7 days. Media was changed every other day in both assays. The same antibodies at the same concentrations were used as described in the tissue sections above, and Images were acquired on an Echo Revolve microscope. For Midkine experiments, passaged primary endothelial and mural cells at passage 3–7 were added at 20,000 cells per well and incubuated with recombinant Midkine (20 ng/ml) or Midkine inhibitor iMDK (100 nM). Images were acquired at 3, 6, 12, and 24 hours on an ECHO REVOLVE microscope and branch points manually quantified in ImageJ.
Single-cell RNA sequencing. Endothelial and mural cells from 7 cases, GW 15–23, were utilized in these experiments (see table under human tissue samples). Following a similar protocol to our previous work (Bhaduri et al., 2020), single-cell capture from live cells was performed following the 10x v2 and v3 Chromium manufacturer’s instructions. In each sample, 10,000 cells were targeted for capture and 12 cycles of amplification for each of the cDNA amplification and library amplification were performed. Libraries were sequenced as per manufacturer recommendation on a NovaSeq S2 flow cell.
Clustering.
We first explored the cell-type identities of endothelial and mural cells using Louvain-Jaccard clustering (Nowakowski et al., 2017). Prior to clustering, batch correction was performed in a similar way to previous approaches (Peng et al., 2019). Each set of cells within a batch was normalized to the highest expressing gene, making the range of expression from 0 to 1. These values were multiplied by the average number of counts within the batch. These normalized datasets were piped into Seurat v.2 (Butler et al., 2018), and in the case of individual sample analysis (such as GW23 mural and/or endothelial cells) log-normalized counts were used. After transfer to Seurat, cells with fewer than 200 genes per cell or more than 10% of reads aligning to mitochondrial genes were discarded. Normalized counts matrices were log2-transformed, and variable genes were calculated using default Seurat parameters. Data were scaled in the space of these variables, and the batch was regressed out. Principal component analysis was performed using FastPCA, and significant principal components were identified using a published formula (Shekhar et al., 2016). In the space of these significant principal components, the k = 10 nearest neighbors were identified as per the RANN R package. The distances between these neighbors were weighted by their Jaccard distance, and Louvain clustering was performed using the igraph R package. If any clusters contained only one cell, the process was repeated with k = 11 and upwards until no clusters contained only one cell. Cluster markers were set to have a minimum percentage of 25% in either population in addition to Seurat package default parameters. UMAPs were generated with Seurat default parameters.
RNA velocity.
Velocity estimates were calculated using the veloctyo.py and scvelo algorithms, as previously described (Eze et al., 2021). Reads that passed quality control after clustering were used as input for the velocyto command line implementation. The human expressed repeat annotation file was retrieved from the UCSC Genome Browser. The genome annotation file used was provided via CellRanger. The output loom file was used as input to estimate velocity through scvelo. For each individual analysis, cells were filtered based on the following parameters: minimum total counts ≥ 200, minimum spliced counts ≥ 20 and minimum unspliced counts ≥ 10. For the combined cortical analysis, the processed loom files for each individual analysis were combined to generate a new UMI count matrix of 15,473 genes across 53,096 cells, for which the velocity embedding was estimated using the stochastic model. For the combined progenitor analysis, cells that were identified as progenitors were used to create the loom file. The loom files for each of the individuals were combined for a total count matrix of 14,207 genes across 30,562 embedding using the same criteria. Each embedding was visualized using UMAP.
RNAscope.
RNAscope experiments were performed according to their protocol for integrated Co-detection for fixed frozen tissue and the Multiplex fluorescent v2 Assay. In brief, slides were first baked at 60°C for 30 minutes, followed by a 15 minute fixation in 4% paraformaldehyde. We then performed target retrieval for 5 minutes using the RNAscope solution in an Oster steamer as recommended by ACD. After target retrieval, we dipped the slides in water, then 100% ethanol, and finally clipped the slides to the rack and drew a hydrophobic barrier. Once the barrier was dry, we added the protease plus solution for 30 minutes in the RNAscope Hybridization oven at 40°C for 30 minutes. During this incubation, we warmed the probes for 10 minutes in a 40°C water bath and then allowed them to cool for 10 minutes. Following the protease plus incubation, we washed the slides in distilled water twice, and then added the probes for 2 hours in the RNAscope Hybridization oven at 40°C. After the probe hybridization, we added the Amp 1 and 2 solutions successively for 30 minutes in the RNAscope Hybridization oven at 40°C and then Amp 3 solution for 15 minutes. All of these steps were interspersed with 2 washes for 2 minutes in the ACD wash buffer solution. We then added the HRP-C1 signal for 15 minutes in the RNAscope Hybridization oven at 40°C, followed by the diluted Opal Dye for signal development for 30 minutes in the RNAscope Hybridization oven at 40°C, and finally a 15 minute incubation in the RNAscope Hybridization oven at 40°C with the HRP blocker. In between all of these steps, we again performed 2 washes for 2 minutes with the ACD wash buffer solution. We then repeated the HRP signal, the Opal Dye, and the HRP blocker for the C2 probes. We then transitioned into our immunohistochemistry protocol listed earlier in this section and blocked for one hour with TBS++++. We then added the primary antibody overnight in TBS++++ and performed secondary staining the following day. Of note, the target retrieval step for RNAscope significantly decreased the sensitivity of antibody staining and we added antibodies 10x more concentrated than for immunohistochemistry alone per ACD guidelines. Due to the postmortem collection of our samples, they are not perfused and have red blood cells in the vasculature. These red blood cells generate significant autofluorescence with the RNAscope workflow and were removed from the images with post-image processing in ImageJ. Raw images available upon request.
CellChat.
Cellchat analysis was done as described in (Jin et al., 2021). Default ‘trimean’ threshold was used for more stringent identification of interactions. Minimum cell number per group was set to 10. Plots were generated as described in Jin et al. 2021 with modifications as noted in our code which is provided on SRA.
Seahorse experiments.
Respiration and glycolytic rates were measured in endothelial cells using a 96-well Seahorse XF96 Extracellular Flux Analyzer. Endothelial cells were plated in XF96 microplates (20,000 cells per well) and incubated overnight. A Seahorse 96-well assay cartridge was placed in a CO2-free incubator with calibration medium overnight. On the day of the assay, cells were incubated with Seahorse XF DMEM Medium, supplemented with 10 mM glucose, 5 mM sodium pyruvate, and 2 mM glutamine, pH = 7.4, and incubated in a CO2-free incubator for 1 h. Oxygen consumption rate (OCR) was measured at baseline and after the addition of respiratory inhibitor FCCP (4μM carbonyl cyanide-4 (trifluoromethoxy) phenylhydrazone, which uncouples oxidative phosphorylation from membrane potential), followed by rotenone (2μM a complex I inhibitor). Extracellular acidification rate (ECAR) was measured at baseline and after the addition of respiratory inhibitor oligomycin A (5μM an ATP synthase inhibitor), followed by rotenone. After each assay was completed, cells were fixed with 4% paraformaldehyde, and the OCR and ECAR signals were normalized to the number of cells in each well, estimated using Hoescht 33342 staining, as well as to the baseline reading at the initial timepoint.
Organoid transplant experiments.
Organoid cultures were grown as previously described (27). Endothelial and mural cells from 6 cases, GW 13–23, were utilized in these experiments (see table under human tissue samples). NIH 3T3 cells were cultured in DMEM/High glucose media (Cytiva) as recommended and transplanted at passage 2. Immediately following FACS experiments, endothelial and mural cells were infected with AAV-CMV-GFP (Vector Biolabs) at 1:2000 with an approximate infection ratio of 25% and incubated at 37°C for 30 minutes. The cells were then washed twice with 1X PBS and gently added on top of 8–10 week old organoids. Organoids were stabilized on Millicell hanging culture inserts (Sigma) and incubated at 37°C with the transplanted cells for 30 minutes before being immersed in media. Media was changed three times per week and transplanted cells were cultured for 2 weeks before being fixed with PFA. Fixed organoids were embedded in an equal parts mix of 30% Sucrose and OCT and sectioned for immunostaining. We performed immunohistochemistry as described above. GFP-positive organoid regions were defined as being within 350μm from multiple GFP(+) cells. GFP-negative organoid regions were defined as being greater than 350μm from any GFP(+) cells.
Cell browser.
The interactive viewer at https://vascular-dev.cells.ucsc.edu was created with the tools from the UCSC Cell Browser package (https://urldefense.proofpoint.com/v2/url?u=https-3Agithub.com_maximilianh_cellBrowser&d=DwIFaQ&c=iORugZls2LlYyCAZRB3XLg&r=gDfG4LoneiL92oJnZQfO8eN8b_paD72mG14zH9dq9HA&m=4YISTHA_IFiTyFUpiw_spcf9rZQpCSYEJl8Q4Db72Uw&s=V6WLAOUJqEo1EORxKrIJFOQ15xjqqSzzCDZz13MXJ0w&e=). Data was exported from the Seurat object with cbImportSeurat, marker genes were manually added and annotated with cbMarkerAnnotate, and configuration files adapted based on information in the manuscript.
Quantification and Statistical Analysis
GraphPad Prism (version 8.4.3) was used to generate all graphs. Comparisons of two variables were determined with Student’s t-tests. Throughout the paper, * indicates p<0.05, ** indicates p<0.01, *** indicates p<0.005, **** indicates p<0.001. The details of what n represents are defined in each figure legend. Non-statistically significant comparisons are not shown. ANOVAs were used for comparisons with multiple groups with post-hoc t-tests to assess differences within groups. The assumptions of the ANOVA were checked using Q-Q plots to assess normality and a plot of residual vs. predicted was used to assess constant variance. Data are represented as mean ± S.E.M.
Supplementary Material
Movie S3. Imaris 3D rendering of RNAscope for ATP1A2 (magenta), KCNJ8 (green), and immunostaining for PDGFR-β (white) demonstrating ATP1A2+KCNJ8+ cells with a typical bulbous classic pericyte morphology sitting on top of endothelial cells at 15 GW, related to Figure 4.
Figure S1. Angiogenic zone in the pallial periventricular region and cerebral cortex in the prenatal human brain, related to Figure 1 A. Low power view of the periventricular area of the developing human brain immunostained with CD31, NG2 and PDGFR-β at 39 GW (top panels) and higher power images of the boxed areas in the ventricular zone (VZ).
B. Immunostaining of CD31, NG2 and PDGFR-β in representative sections of the human pallial periventricular region at 14 and 22 GW.
C-D. Quantification of CD31+ and NG2+ surface area in the pallial VZ.
E. Immunostaining of CD31, NG2 and PDGFR-β in the human cerebral cortex at 17 GW.
F. Quantification of CD31+ surface area in the human cortex at 17 GW.
G. Confocal Z stack image of CD34, MKI67 and PDGFR-β in the cerebral cortex at 14 and 21 GW.
H-I. Quantification of the number of MKI67+ endothelial (H) or mural (I) cells in the pallial VZ.
In C, D, F, H, and I, each data point represents the average of 5 sections from one case. ANOVAs were used for comparisons with multiple groups with post-hoc Student’s t-tests to assess differences within groups. Statistics shown represent the post-hoc t-tests, and non-significant comparisons not shown. Data are represented as mean ± S.E.M. and non-significant comparisons not shown.
Figure S3. Flow cytometry-based isolation of vascular cells from the prenatal human brain, related to Figure 3 A. Sequential gating and purification of myeloid cells (CD45+CD31−ANPEP−), endothelial cells (CD45−CD31+ANPEP−), and mural cells (CD45−CD31−ANPEP+) from the prenatal human brain. Note that the majority of CD45+ cells are also CX3CR1+, supporting their identity as myeloid cells.
B. Additional immunostainings for vascular markers with FACS showed that all CD31+ cells were also CD34+, CD105lo, CD146+, and PDGFR-β−. In contrast, mural cells were PDGFR-β+, CD34−, CD146high and CD105lo.
C. FACS plots of controls with secondary antibodies only.
D. Violin plots for the expression of markers for other cell types, including radial glia (SOX2), neuroblasts (DCX), microglia (AIF1), and oligodendrocytes (OLIG2) compared to CD34 and PDGFR-β.
Figure S4. Subtype- and age-dependent gene ontology features of endothelial cells defined by single-cell transcriptomics, related to Figure 4 A. Gene ontology (GO) groups represented by each subtype of endothelial cells color-coded by GO terms with different patterns of relatedness between the endothelial subtypes.
B. Venn diagrams showing overlapping marker gene expression between P7 mouse endothelial cells (from Sabbagh et al) and second trimester human endothelial cells (this study).
C. Feature plots for genes enriched in proliferative endothelial cells (TOP2A, CENPF, AURKB, and MKI67), arterial endothelial cells (FBLN5, CXCL12, GJA4), tip cells (ADM, PGF, ANGPT2), and capillary/venous endothelial cells (ADGRG6, MFSD2A) in RNA velocity UMAP space at 15 GW (top panel) and 23 GW (bottom panel).
D-E. Quantification of mitochondrial surface area with confocal microscopy. D. Representative confocal z stack image of 17 GW MGE stained for TOMM20 (mitochondria) and CD34 (endothelial cells). E. Quantification of mitochondrial surface area in early versus late second trimester samples. In E, each data point represents a separate case. Student’s t-test was used to assess differences between groups. Data are represented as mean ± S.E.M.
Figure S5. Subtype- and age-dependent gene ontology features of mural cells defined by single-cell transcriptomics, related to Figure 5 A. Gene ontology (GO) groups represented by each subtype of mural cells color-coded by GO terms with different patterns of relatedness between the mural subtypes.
B. Venn diagrams showing overlapping marker gene expression between adult mouse mural cells (from Vanlandewijck et al) and second trimester human endothelial cells (this study).
C. Volcano plot of differentially expressed genes in young (left side) versus old (right side).
D. Gene ontology groups defined by genes enriched in young (left side) and old (right side) mural cells.
E. Feature plots for genes enriched in proliferative mural cells (UBE2C, BIRC5, and MKI67), smooth muscle cells (MYL9, TAGLN), fibroblasts (SERPING1, LUM), and capillary/venous cells (ATP1A2, KCNJ8) in RNA velocity UMAP space at 15 GW (top panel) and 23 GW (bottom panel).
Figure S6. Molecular pathways critical for the cell-cell communication between endothelial and mural cells in the prenatal human brain, related to Figure 6 A-F. Chord and violin plots for genes in the Collagen (A), Laminin (B), Occludin (C), Notch (D), Midkine (E), and VEGF families (F) at 15 and 23GW in all vascular subtypes.
Figure S2. Ultrastructural characteristics of developing human MGE and cortex vasculature, related to Figure 2.
A. Micrographs of ultrathin sections from the 23 GW hMGE showing vascular distribution (magenta). hMGE Blood vessels located in the outer SVZ (oSVZ) were associated with bundles of radial glial expansions (blue).
B-C. Blood vessel located in the 23 GW MGE VZ composed of intermediate (blue) and light (purple) endothelial cells and a mural cell (magenta). Note the very thin vascular lumen (magenta line). Junctional complexes indicated by double arrows.
D. Serial sections of a blood vessel composed of light (EL) and intermediate (Ei) endothelial cells with very thin vascular lumen (magenta) and discontinuous basal lamina (yellow).
E. Serial sections of mural (magenta) and all three types of endothelial cells engaging in extensive interdigitations.
F. Micrographs of ultrathin sections from the 21 GW cortex showing vascular distribution (magenta).
G-I. High magnification of a blood vessel from the cerebral cortex showing extensive junctions between the three types of endothelial cells. Junctional complexes indicated by double arrows. Scale bars: G: (10 μm), A and F: (5 μm), B, E and H: (2 μm), C, D and I: (500nm).
J. Quantification of luminal area in blood vessels at different GW and zones. 20 blood vessels quantified in each zone and age.
Figure S7 Human endothelial and mural cells transplanted into cortical organoids promote neurogenesis and reduce cell stress, related to Figure 7. A. Confocal images for immunostains for GFP, SATB2, and PAX6 in the transplanted organoids.
B. Confocal images for immunostains for GFP and CUX1/2 in the transplanted organoids.
C-E. Quantification of SATB2+ (C), PAX6+ (D), and CUX1/2+ (E) cells in organoids in areas with GFP+ (transplanted) or control regions.
F-G. Confocal images for immunostains for GFP and Enolase (F) or GORASP2 (G) in the transplanted organoids.
H-I. Quantification of Enolase+ (H) or GORASP2+ (I) cells in organoids in areas with GFP+ (transplanted) or control regions.
J. A working model illustrating that diverse endothelial and mural cells contribute to blood vessel development in the cerebral cortex and germinal matrix of the prenatal human brain. A schematic diagram of the prenatal human brain shows an ensemble of endothelial and mural cells can be identified in the ventricular zone in the ganglionic eminences and the cerebral cortex. Both tip cells and proliferative endothelial cells are located close to the ventricular zone, but not in the cerebral cortex, whereas other endothelial cell subtypes, including capillary, venous and arterial cells are found to tile the nascent vasculature in both germinal matrix and cerebral cortex in a mosaic pattern. Proliferative mural cells are also found primarily close to the ventricular zone in the ganglionic eminences. Classic pericytes and smooth muscle cells are identified in both the germinal matrix and the cerebral cortex, whereas the fibroblasts are more abundant in the cerebral cortex.
In C-E, H and I, each data point represents the average of 5 sections from one transplant. Student’s t-tests were used to assess differences within groups. Data are represented as mean ± S.E.M. and non-significant comparisons not shown. Control regions were defined as >350 μm from GFP+ transplanted cells.
Movie S2. Imaris 3D rendering of Ki-67+ (magenta) endothelial cell (CD31+, green) in the 17 GW GE showing proliferative endothelial cells in a complex vascular structure close to the ventricular zone, related to Figures 1 and S1.
Movie S1. Imaris 3D rendering of CD31 (green) and NG2 (blue) showing prominent filopodia next to the ventricular zone, related to Figure 1.
Table S2 Metadata and sequencing depth, related to Figure 3.
Table S3 Subtype markers for endothelial and mural cells, related to Figures 3, 4 and S3, and S4.
Table S4 Differentially expressed genes by younger or older endothelial or mural cells, related to Figures 3, 4 and S3, and S4.
Highlights.
Diverse vascular cell subtypes tile the nascent vasculature in prenatal human brain
Vascular cells exhibit stage-specific developmental trajectories and bioenergetics
Midkine-mediated signaling promotes morphogenesis of endothelial and mural cells
Vascularization of brain organoids promote neurogenesis and reduce cellular stress
Acknowledgements.
We are grateful to members of the Crouch, Kriegstein, and Huang labs for their expertise and critical discussion. We also appreciate the statistical support from Dr. Ann Lazar from the UCSF Clinical Translational Science Institute. The authors acknowledge the Biological Imaging Development CoLabs for training and use of their equipment and software.
Funding
This study has been supported by the Eli and Edythe Broad Regeneration Medicine and Stem Cell Fellowship, Pediatric Scientist Development Program (5K12HD000850-34), UCSF Physician Scientist Scholars Program (PSSP), American Heart Association Career Development Grant 857876, CIRM Alpha Stem Cell Clinic Fellowship, and NIH K08 NS116161 to E.E.C., NIH K99 NS111731 to A.B., NIH F31 F31NS117121 to U.C.E., NIH K99 MH125329 to M.G.A., California Institute of Regenerative Medicine (CIRM) Bridges Science Master’s Fellowship (SFSU EDUC2-12693) to E.J.V., CIRM Regenerative Medicine Research Training Program to N.K.B., AHA Predoctoral Fellowship 19PRE3480616 and UCSF Discovery Fellowship to J.C., NIH P01 NS083513 Diversity Supplement to C. M., NHGRI 5U41HG002371-19 to M.H., NIH K08 NS091537 to M.F.P., R01 AG065428 to K.N., P01 NS083513 and U01 MH105989 to E.J.H., A.A-B., and A.R.K. The UCSF Flow Cytometry Core is supported by DRC Center Grant NIH P30 DK063720.
Inclusions and diversity statement
We ensured gender balance in the recruitment of human subjects. One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science, a member of the LGBTQ+ community, or received support from a program designed to promote minority representation in science. While citing references scientifically relevant for this work, we also actively worked to promote gender balance in our reference list.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests A.R.K. and A.A-B. are founding members of Neurona Therapeutics.
Bibliography
- Ando K, Tong L, Peng D, Vázquez-Liébanas E, Chiyoda H, He L, Liu J, Kawakami K, Mochizuki N, Fukuhara S, et al. (2022). KCNJ8/ABCC9-containing K-ATP channel modulates brain vascular smooth muscle development and neurovascular coupling. Dev. Cell [DOI] [PubMed] [Google Scholar]
- Armulik A, Genové G, Mäe M, Nisancioglu MH, Wallgard E, Niaudet C, He L, Norlin J, Lindblom P, Strittmatter K, et al. (2010). Pericytes regulate the blood-brain barrier. Nature 468, 557–561. [DOI] [PubMed] [Google Scholar]
- Armulik A, Genové G, and Betsholtz C (2011). Pericytes: developmental, physiological, and pathological perspectives, problems, and promises. Dev. Cell 21, 193–215. [DOI] [PubMed] [Google Scholar]
- Attwell D, Mishra A, Hall CN, O’Farrell FM, and Dalkara T (2016). What is a pericyte? J. Cereb. Blood Flow Metab 36, 451–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayloo S, Lazo CG, Sun S, Zhang W, Cui B, and Gu C (2022). Pericyte-to-endothelial cell signaling via vitronectin-integrin regulates blood-CNS barrier. Neuron 110, 1641–1655.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaduri A, Andrews MG, Mancia Leon W, Jung D, Shin D, Allen D, Jung D, Schmunk G, Haeussler M, Salma J, et al. (2020). Cell stress in cortical organoids impairs molecular subtype specification. Nature 578, 142–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Korte N, Nortley R, Sethi H, Tang Y, and Attwell D (2018). Targeting pericytes for therapeutic approaches to neurological disorders. Acta Neuropathol. 136, 507–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossutta M, Darche M, Carpentier G, Houppe C, Ponzo M, Raineri F, Vallée B, Gilles M-E, Villain D, Picard E, et al. (2019). Weibel-Palade Bodies Orchestrate Pericytes During Angiogenesis. Arterioscler. Thromb. Vasc. Biol 39, 1843–1858. [DOI] [PubMed] [Google Scholar]
- Crouch EE, and Doetsch F (2018). FACS isolation of endothelial cells and pericytes from mouse brain microregions. Nat. Protoc 13, 738–751. [DOI] [PubMed] [Google Scholar]
- Crouch EE, Liu C, Silva-Vargas V, and Doetsch F (2015). Regional and stage-specific effects of prospectively purified vascular cells on the adult V-SVZ neural stem cell lineage. J. Neurosci 35, 4528–4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daneman R, Zhou L, Kebede AA, and Barres BA (2010). Pericytes are required for blood-brain barrier integrity during embryogenesis. Nature 468, 562–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Palma C, Falcone S, Panzeri C, Radice S, Bassi MT, and Clementi E (2008). Endothelial nitric oxide synthase overexpression by neuronal cells in neurodegeneration: a link between inflammation and neuroprotection. J. Neurochem 106, 193–204. [DOI] [PubMed] [Google Scholar]
- Di Marco B, Crouch EE, Shah B, Duman C, Paredes MF, Ruiz de Almodovar C, Huang EJ, and Alfonso J (2020). Reciprocal Interaction between Vascular Filopodia and Neural Stem Cells Shapes Neurogenesis in the Ventral Telencephalon. Cell Rep. 33, 108256. [DOI] [PubMed] [Google Scholar]
- Duan L, Zhang X-D, Miao W-Y, Sun Y-J, Xiong G, Wu Q, Li G, Yang P, Yu H, Li H, et al. (2018). PDGFRβ Cells Rapidly Relay Inflammatory Signal from the Circulatory System to Neurons via Chemokine CCL2. Neuron 100, 183–200.e8. [DOI] [PubMed] [Google Scholar]
- Ehling M, Adams S, Benedito R, and Adams RH (2013). Notch controls retinal blood vessel maturation and quiescence. Development 140, 3051–3061. [DOI] [PubMed] [Google Scholar]
- Eze UC, Bhaduri A, Haeussler M, Nowakowski TJ, and Kriegstein AR (2021). Single-cell atlas of early human brain development highlights heterogeneity of human neuroepithelial cells and early radial glia. Nat. Neurosci 24, 584–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faraci FM (2006). Protecting the Brain With eNOS. Circ. Res 99, 1029–1030. [DOI] [PubMed] [Google Scholar]
- Fernández-Chacón M, García-González I, Mühleder S, and Benedito R (2021). Role of Notch in endothelial biology. Angiogenesis 24, 237–250. [DOI] [PubMed] [Google Scholar]
- Filippou PS, Karagiannis GS, and Constantinidou A (2020). Midkine (MDK) growth factor: a key player in cancer progression and a promising therapeutic target. Oncogene 39, 2040–2054. [DOI] [PubMed] [Google Scholar]
- Fu Y, Yang M, Yu H, Wang Y, Wu X, Yong J, Mao Y, Cui Y, Fan X, Wen L, et al. (2021). Heterogeneity of glial progenitor cells during the neurogenesis-to-gliogenesis switch in the developing human cerebral cortex. Cell Rep. 34, 108788. [DOI] [PubMed] [Google Scholar]
- Garcia FJ, Sun N, Lee H, Godlewski B, Mathys H, Galani K, Zhou B, Jiang X, Ng AP, Mantero J, et al. (2022). Single-cell dissection of the human brain vasculature. Nature 603, 893–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gee HY, Noh SH, Tang BL, Kim KH, and Lee MG (2011). Rescue of ΔF508-CFTR trafficking via a GRASP-dependent unconventional secretion pathway. Cell 146, 746–760. [DOI] [PubMed] [Google Scholar]
- Gould DB, Phalan FC, Breedveld GJ, van Mil SE, Smith RS, Schimenti JC, Aguglia U, van der Knaap MS, Heutink P, and John SWM (2005). Mutations in Col4a1 cause perinatal cerebral hemorrhage and porencephaly. Science 308, 1167–1171. [DOI] [PubMed] [Google Scholar]
- Hall CN, Reynell C, Gesslein B, Hamilton NB, Mishra A, Sutherland BA, O’Farrell FM, Buchan AM, Lauritzen M, and Attwell D (2014). Capillary pericytes regulate cerebral blood flow in health and disease. Nature 508, 55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan SS, Tsaryk R, Lange M, Wisniewski L, Moore JC, Lawson ND, Wojciechowska K, Schnittler H, and Siekmann AF (2017). Endothelial Notch signalling limits angiogenesis via control of artery formation. Nat. Cell Biol 19, 928–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan KA, Ambler CA, Chapman DL, and Bautch VL (2004). The neural tube patterns vessels developmentally using the VEGF signaling pathway. Development 131, 1503–1513. [DOI] [PubMed] [Google Scholar]
- James JM, Gewolb C, and Bautch VL (2009). Neurovascular development uses VEGF-A signaling to regulate blood vessel ingression into the neural tube. Development 136, 833–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S, Guerrero-Juarez CF, Zhang L, Chang I, Ramos R, Kuan C-H, Myung P, Plikus MV, and Nie Q (2021). Inference and analysis of cell-cell communication using CellChat. Nat. Commun 12, 1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung B, Arnold TD, Raschperger E, Gaengel K, and Betsholtz C (2018). Visualization of vascular mural cells in developing brain using genetically labeled transgenic reporter mice. J. Cereb. Blood Flow Metab 38, 456–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Noh SH, Piao H, Kim DH, Kim K, Cha JS, Chung WY, Cho H-S, Kim JY, and Lee MG (2016). Monomerization and ER relocalization of GRASP is a requisite for unconventional secretion of CFTR. Traffic 17, 733–753. [DOI] [PubMed] [Google Scholar]
- Kisler K, Nelson AR, Rege SV, Ramanathan A, Wang Y, Ahuja A, Lazic D, Tsai PS, Zhao Z, Zhou Y, et al. (2017). Pericyte degeneration leads to neurovascular uncoupling and limits oxygen supply to brain. Nat. Neurosci 20, 406–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komabayashi-Suzuki M, Yamanishi E, Watanabe C, Okamura M, Tabata H, Iwai R, Ajioka I, Matsushita J, Kidoya H, Takakura N, et al. (2019). Spatiotemporally Dependent Vascularization Is Differently Utilized among Neural Progenitor Subtypes during Neocortical Development. Cell Rep. 29, 1113–1129.e5. [DOI] [PubMed] [Google Scholar]
- La Manno G, Gyllborg D, Codeluppi S, Nishimura K, Salto C, Zeisel A, Borm LE, Stott SRW, Toledo EM, Villaescusa JC, et al. (2016). Molecular diversity of midbrain development in mouse, human, and stem cells. Cell 167, 566–580.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, Lidschreiber K, Kastriti ME, Lönnerberg P, Furlan A, et al. (2018). RNA velocity of single cells. Nature 560, 494–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacar B, Herman P, Platel J-C, Kubera C, Hyder F, and Bordey A (2012). Neural progenitor cells regulate capillary blood flow in the postnatal subventricular zone. J. Neurosci 32, 16435–16448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Sun X, and Carmeliet P (2019). Hallmarks of endothelial cell metabolism in health and disease. Cell Metab. 30, 414–433. [DOI] [PubMed] [Google Scholar]
- Licht T, and Keshet E (2015). The vascular niche in adult neurogenesis. Mech. Dev 138 Pt 1, 56–62. [DOI] [PubMed] [Google Scholar]
- Nowakowski TJ, Bhaduri A, Pollen AA, Alvarado B, Mostajo-Radji MA, Di Lullo E, Haeussler M, Sandoval-Espinosa C, Liu SJ, Velmeshev D, et al. (2017). Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science 358, 1318–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brown NM, Megason SG, and Gu C (2019). Suppression of transcytosis regulates zebrafish blood-brain barrier function. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredes MF, James D, Gil-Perotin S, Kim H, Cotter JA, Ng C, Sandoval K, Rowitch DH, Xu D, McQuillen PS, et al. (2016). Extensive migration of young neurons into the infant human frontal lobe. Science 354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredes MF, Mora C, Flores-Ramirez Q, Cebrian-Silla A, Del Dosso A, Larimer P, Chen J, Kang G, Gonzalez Granero S, Garcia E, et al. (2022). Nests of dividing neuroblasts sustain interneuron production for the developing human brain. Science 375, eabk2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker KR, Migliorini D, Perkey E, Yost KE, Bhaduri A, Bagga P, Haris M, Wilson NE, Liu F, Gabunia K, et al. (2020). Single-Cell Analyses Identify Brain Mural Cells Expressing CD19 as Potential Off-Tumor Targets for CAR-T Immunotherapies. Cell 183, 126–142.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y-R, Shekhar K, Yan W, Herrmann D, Sappington A, Bryman GS, van Zyl T, Do MTH, Regev A, and Sanes JR (2019). Molecular classification and comparative taxonomics of foveal and peripheral cells in primate retina. Cell 176, 1222–1237.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitulescu ME, Schmidt I, Giaimo BD, Antoine T, Berkenfeld F, Ferrante F, Park H, Ehling M, Biljes D, Rocha SF, et al. (2017). Dll4 and Notch signalling couples sprouting angiogenesis and artery formation. Nat. Cell Biol 19, 915–927. [DOI] [PubMed] [Google Scholar]
- Polioudakis D, de la Torre-Ubieta L, Langerman J, Elkins AG, Shi X, Stein JL, Vuong CK, Nichterwitz S, Gevorgian M, Opland CK, et al. (2019). A Single-Cell Transcriptomic Atlas of Human Neocortical Development during Mid-gestation. Neuron 103, 785–801.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollen AA, Nowakowski TJ, Chen J, Retallack H, Sandoval-Espinosa C, Nicholas CR, Shuga J, Liu SJ, Oldham MC, Diaz A, et al. (2015). Molecular identity of human outer radial glia during cortical development. Cell 163, 55–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohlenova K, Goveia J, García-Caballero M, Subramanian A, Kalucka J, Treps L, Falkenberg KD, de Rooij LPMH, Zheng Y, Lin L, et al. (2020). Single-Cell RNA Sequencing Maps Endothelial Metabolic Plasticity in Pathological Angiogenesis. Cell Metab. 31, 862–877.e14. [DOI] [PubMed] [Google Scholar]
- Ross-Munro E, Kwa F, Kreiner J, Khore M, Miller SL, Tolcos M, Fleiss B, and Walker DW (2020). Midkine: the who, what, where, and when of a promising neurotrophic therapy for perinatal brain injury. Front. Neurol 11, 568814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbagh MF, Heng JS, Luo C, Castanon RG, Nery JR, Rattner A, Goff LA, Ecker JR, and Nathans J (2018). Transcriptional and epigenomic landscapes of CNS and non-CNS vascular endothelial cells. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekhar K, Lapan SW, Whitney IE, Tran NM, Macosko EZ, Kowalczyk M, Adiconis X, Levin JZ, Nemesh J, Goldman M, et al. (2016). Comprehensive Classification of Retinal Bipolar Neurons by Single-Cell Transcriptomics. Cell 166, 1308–1323.e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Vargas V, Crouch EE, and Doetsch F (2013). Adult neural stem cells and their niche: a dynamic duo during homeostasis, regeneration, and aging. Curr. Opin. Neurobiol 23, 935–942. [DOI] [PubMed] [Google Scholar]
- Smith RS, Florio M, Akula SK, Neil JE, Wang Y, Hill RS, Goldman M, Mullally CD, Reed N, Bello-Espinosa L, et al. (2021). Early role for a Na+,K+-ATPase (ATP1A3) in brain development. Proc. Natl. Acad. Sci. USA 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Liu WA, Zhang X-J, Shi W, Ren S-Q, Li Z, Brown KN, and Shi S-H (2016). Vascular influence on ventral telencephalic progenitors and neocortical interneuron production. Dev. Cell 36, 624–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlandewijck M, He L, Mäe MA, Andrae J, Ando K, Del Gaudio F, Nahar K, Lebouvier T, Laviña B, Gouveia L, et al. (2018). A molecular atlas of cell types and zonation in the brain vasculature. Nature 554, 475–480. [DOI] [PubMed] [Google Scholar]
- Volz KS, Jacobs AH, Chen HI, Poduri A, McKay AS, Riordan DP, Kofler N, Kitajewski J, Weissman I, and Red-Horse K (2015). Pericytes are progenitors for coronary artery smooth muscle. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Dai X, Wu S, Xu W, Song P, and Huang K (2021). FUNDC1-dependent mitochondria-associated endoplasmic reticulum membranes are involved in angiogenesis and neoangiogenesis. Nat. Commun 12, 2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weckbach LT, Groesser L, Borgolte J, Pagel J-I, Pogoda F, Schymeinsky J, Müller-Höcker J, Shakibaei M, Muramatsu T, Deindl E, et al. (2012). Midkine acts as proangiogenic cytokine in hypoxia-induced angiogenesis. Am. J. Physiol. Heart Circ. Physiol 303, H429–38. [DOI] [PubMed] [Google Scholar]
- Winkler EA, Kim CN, Ross JM, Garcia JH, Gil E, Oh I, Chen LQ, Wu D, Catapano JS, Raygor K, et al. (2022). A single-cell atlas of the normal and malformed human brain vasculature. Science 375, eabi7377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Hasan SS, Schmidt I, Rocha SF, Pitulescu ME, Bussmann J, Meyen D, Raz E, Adams RH, and Siekmann AF (2014). Arteries are formed by vein-derived endothelial tip cells. Nat. Commun 5, 5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S, Muramatsu M, Azuma E, Ikutani M, Nagai Y, Sagara H, Koo B-N, Kita S, O’Donnell E, Osawa T, et al. (2017). A subset of cerebrovascular pericytes originates from mature macrophages in the very early phase of vascular development in CNS. Sci. Rep 7, 3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang AC, Vest RT, Kern F, Lee DP, Agam M, Maat CA, Losada PM, Chen MB, Schaum N, Khoury N, et al. (2022). A human brain vascular atlas reveals diverse mediators of Alzheimer’s risk. Nature 603, 885–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S3. Imaris 3D rendering of RNAscope for ATP1A2 (magenta), KCNJ8 (green), and immunostaining for PDGFR-β (white) demonstrating ATP1A2+KCNJ8+ cells with a typical bulbous classic pericyte morphology sitting on top of endothelial cells at 15 GW, related to Figure 4.
Figure S1. Angiogenic zone in the pallial periventricular region and cerebral cortex in the prenatal human brain, related to Figure 1 A. Low power view of the periventricular area of the developing human brain immunostained with CD31, NG2 and PDGFR-β at 39 GW (top panels) and higher power images of the boxed areas in the ventricular zone (VZ).
B. Immunostaining of CD31, NG2 and PDGFR-β in representative sections of the human pallial periventricular region at 14 and 22 GW.
C-D. Quantification of CD31+ and NG2+ surface area in the pallial VZ.
E. Immunostaining of CD31, NG2 and PDGFR-β in the human cerebral cortex at 17 GW.
F. Quantification of CD31+ surface area in the human cortex at 17 GW.
G. Confocal Z stack image of CD34, MKI67 and PDGFR-β in the cerebral cortex at 14 and 21 GW.
H-I. Quantification of the number of MKI67+ endothelial (H) or mural (I) cells in the pallial VZ.
In C, D, F, H, and I, each data point represents the average of 5 sections from one case. ANOVAs were used for comparisons with multiple groups with post-hoc Student’s t-tests to assess differences within groups. Statistics shown represent the post-hoc t-tests, and non-significant comparisons not shown. Data are represented as mean ± S.E.M. and non-significant comparisons not shown.
Figure S3. Flow cytometry-based isolation of vascular cells from the prenatal human brain, related to Figure 3 A. Sequential gating and purification of myeloid cells (CD45+CD31−ANPEP−), endothelial cells (CD45−CD31+ANPEP−), and mural cells (CD45−CD31−ANPEP+) from the prenatal human brain. Note that the majority of CD45+ cells are also CX3CR1+, supporting their identity as myeloid cells.
B. Additional immunostainings for vascular markers with FACS showed that all CD31+ cells were also CD34+, CD105lo, CD146+, and PDGFR-β−. In contrast, mural cells were PDGFR-β+, CD34−, CD146high and CD105lo.
C. FACS plots of controls with secondary antibodies only.
D. Violin plots for the expression of markers for other cell types, including radial glia (SOX2), neuroblasts (DCX), microglia (AIF1), and oligodendrocytes (OLIG2) compared to CD34 and PDGFR-β.
Figure S4. Subtype- and age-dependent gene ontology features of endothelial cells defined by single-cell transcriptomics, related to Figure 4 A. Gene ontology (GO) groups represented by each subtype of endothelial cells color-coded by GO terms with different patterns of relatedness between the endothelial subtypes.
B. Venn diagrams showing overlapping marker gene expression between P7 mouse endothelial cells (from Sabbagh et al) and second trimester human endothelial cells (this study).
C. Feature plots for genes enriched in proliferative endothelial cells (TOP2A, CENPF, AURKB, and MKI67), arterial endothelial cells (FBLN5, CXCL12, GJA4), tip cells (ADM, PGF, ANGPT2), and capillary/venous endothelial cells (ADGRG6, MFSD2A) in RNA velocity UMAP space at 15 GW (top panel) and 23 GW (bottom panel).
D-E. Quantification of mitochondrial surface area with confocal microscopy. D. Representative confocal z stack image of 17 GW MGE stained for TOMM20 (mitochondria) and CD34 (endothelial cells). E. Quantification of mitochondrial surface area in early versus late second trimester samples. In E, each data point represents a separate case. Student’s t-test was used to assess differences between groups. Data are represented as mean ± S.E.M.
Figure S5. Subtype- and age-dependent gene ontology features of mural cells defined by single-cell transcriptomics, related to Figure 5 A. Gene ontology (GO) groups represented by each subtype of mural cells color-coded by GO terms with different patterns of relatedness between the mural subtypes.
B. Venn diagrams showing overlapping marker gene expression between adult mouse mural cells (from Vanlandewijck et al) and second trimester human endothelial cells (this study).
C. Volcano plot of differentially expressed genes in young (left side) versus old (right side).
D. Gene ontology groups defined by genes enriched in young (left side) and old (right side) mural cells.
E. Feature plots for genes enriched in proliferative mural cells (UBE2C, BIRC5, and MKI67), smooth muscle cells (MYL9, TAGLN), fibroblasts (SERPING1, LUM), and capillary/venous cells (ATP1A2, KCNJ8) in RNA velocity UMAP space at 15 GW (top panel) and 23 GW (bottom panel).
Figure S6. Molecular pathways critical for the cell-cell communication between endothelial and mural cells in the prenatal human brain, related to Figure 6 A-F. Chord and violin plots for genes in the Collagen (A), Laminin (B), Occludin (C), Notch (D), Midkine (E), and VEGF families (F) at 15 and 23GW in all vascular subtypes.
Figure S2. Ultrastructural characteristics of developing human MGE and cortex vasculature, related to Figure 2.
A. Micrographs of ultrathin sections from the 23 GW hMGE showing vascular distribution (magenta). hMGE Blood vessels located in the outer SVZ (oSVZ) were associated with bundles of radial glial expansions (blue).
B-C. Blood vessel located in the 23 GW MGE VZ composed of intermediate (blue) and light (purple) endothelial cells and a mural cell (magenta). Note the very thin vascular lumen (magenta line). Junctional complexes indicated by double arrows.
D. Serial sections of a blood vessel composed of light (EL) and intermediate (Ei) endothelial cells with very thin vascular lumen (magenta) and discontinuous basal lamina (yellow).
E. Serial sections of mural (magenta) and all three types of endothelial cells engaging in extensive interdigitations.
F. Micrographs of ultrathin sections from the 21 GW cortex showing vascular distribution (magenta).
G-I. High magnification of a blood vessel from the cerebral cortex showing extensive junctions between the three types of endothelial cells. Junctional complexes indicated by double arrows. Scale bars: G: (10 μm), A and F: (5 μm), B, E and H: (2 μm), C, D and I: (500nm).
J. Quantification of luminal area in blood vessels at different GW and zones. 20 blood vessels quantified in each zone and age.
Figure S7 Human endothelial and mural cells transplanted into cortical organoids promote neurogenesis and reduce cell stress, related to Figure 7. A. Confocal images for immunostains for GFP, SATB2, and PAX6 in the transplanted organoids.
B. Confocal images for immunostains for GFP and CUX1/2 in the transplanted organoids.
C-E. Quantification of SATB2+ (C), PAX6+ (D), and CUX1/2+ (E) cells in organoids in areas with GFP+ (transplanted) or control regions.
F-G. Confocal images for immunostains for GFP and Enolase (F) or GORASP2 (G) in the transplanted organoids.
H-I. Quantification of Enolase+ (H) or GORASP2+ (I) cells in organoids in areas with GFP+ (transplanted) or control regions.
J. A working model illustrating that diverse endothelial and mural cells contribute to blood vessel development in the cerebral cortex and germinal matrix of the prenatal human brain. A schematic diagram of the prenatal human brain shows an ensemble of endothelial and mural cells can be identified in the ventricular zone in the ganglionic eminences and the cerebral cortex. Both tip cells and proliferative endothelial cells are located close to the ventricular zone, but not in the cerebral cortex, whereas other endothelial cell subtypes, including capillary, venous and arterial cells are found to tile the nascent vasculature in both germinal matrix and cerebral cortex in a mosaic pattern. Proliferative mural cells are also found primarily close to the ventricular zone in the ganglionic eminences. Classic pericytes and smooth muscle cells are identified in both the germinal matrix and the cerebral cortex, whereas the fibroblasts are more abundant in the cerebral cortex.
In C-E, H and I, each data point represents the average of 5 sections from one transplant. Student’s t-tests were used to assess differences within groups. Data are represented as mean ± S.E.M. and non-significant comparisons not shown. Control regions were defined as >350 μm from GFP+ transplanted cells.
Movie S2. Imaris 3D rendering of Ki-67+ (magenta) endothelial cell (CD31+, green) in the 17 GW GE showing proliferative endothelial cells in a complex vascular structure close to the ventricular zone, related to Figures 1 and S1.
Movie S1. Imaris 3D rendering of CD31 (green) and NG2 (blue) showing prominent filopodia next to the ventricular zone, related to Figure 1.
Table S2 Metadata and sequencing depth, related to Figure 3.
Table S3 Subtype markers for endothelial and mural cells, related to Figures 3, 4 and S3, and S4.
Table S4 Differentially expressed genes by younger or older endothelial or mural cells, related to Figures 3, 4 and S3, and S4.
Data Availability Statement
Single-cell RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Any FACS or microscopy data, confocal, electron, or other, reported in this paper will be shared by the lead contact upon request.
No original code was generated in this manuscript. All previously published algorithms are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key resources table
| REAGENT or RESOURCE | SOURCE | RRID IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-human CD131 | Agilent | AB_2892053 |
| Rabbit anti-human NG2 | Millipore | AB_11213678 |
| Goat anti-human PDGFR-β | R&D | AB_355339 |
| Rabbit anti-human Ki-67 | BD Biosciences | AB_396287 |
| Mouse anti-human CD31-Alexa488 | BD Biosciences | AB_647081 |
| Mouse anti-human CD13-APC | BD Biosciences | AB_398624 |
| Mouse anti-human CD45-PeCy7 | BD Biosciences | AB_396854 |
| Mouse anti-human CX3CR1-Alexa647 | BD Biosciences | AB_2869728 |
| Mouse anti-human PDGFR-β-PE | BD Biosciences | AB_397132 |
| Mouse anti-human CD105-PE | BD Biosciences | AB_2033932 |
| Mouse anti-human CD34-PE | BD Biosciences | AB_10563214 |
| IgG2a, K Mouse, Alexa Fluor 488, Clone: eBM2a, Isotype Control | Thermo Fisher | AB_470233 |
| APC Mouse IgG1, k Isotype Control | BD Biosciences | AB_398613 |
| PE Mouse IgG1, k Isotype Control | BD Biosciences | AB_396091 |
| PE-Cy7 Mouse IgG1 k Isotype Control | BD Biosciences | AB_396914 |
| Rabbit anti-human MFSD2A | ThermoFisher | AB_11152332 |
| Rabbit anti-human LUM | ThermoFisher | MA5–29402 |
| Rabbit anti-human GJA4 | Abcam | AB_2861171 |
| Mouse anti-human ADM | Thermo Fisher | AB_2722821 |
| Mouse anti-human smooth muscle actin | Sigma | AB_476701 |
| Goat anti-human COL4A1 | Millipore | AB_92262 |
| Rabbit anti-human SDC2 | Sigma | AB_2678934 |
| Rabbit anti-human ITGB1 | Sigma | AB_2732634 |
| Chicken anti-human GFP | Abcam | AB_300798 |
| Guinea Pig anti-human RBFOX3 (NeuN) | Fisher | AB_11205592 |
| Rat anti-human BCL11B (CTIP2) | Abcam | AB_2064130 |
| Rabbit anti-human PGK1 | Thermo Fisher | AB_2161216 |
| Sheep anti-human CD34 | R&D | AB_10973459 |
| Goat anti-human Midkine | R&D | AB_2143400 |
| Rabbit anti-human ITGA1 | Abcam | Catalog number ab181434 |
| TOMM20 | Abcam | AB_2889972 |
| CUX1/2 | Abcam | AB_941209 |
| SATB2 | Abcam | AB_882455 |
| PAX6 | Abcam | AB_2750924 |
| Enolase | Abcam | AB_2827927 |
| GORASP2 | Protein Tech | AB_2113473 |
| Bacterial and virus strains | ||
| AAV-CMV-GFP | Vector Labs | Catalog number 7117 |
| Chemicals, peptides, and recombinant proteins | ||
| Triton-X | Sigma | Catalog number T9284 |
| Tris-HCL | Corning | Catalog number 46-031-CM |
| Blocking reagent | Perkin Elmer | Catalog number FP1012 |
| BSA | Sigma | Catalog number A7979-50ML |
| Glucose | Sigma | Catalog number G7021-1KG |
| DNase | Worthington | Catalog number LS002139 |
| Collagenase/dispase | Sigma | Catalog number 11097113001 |
| Percoll | Signa | Catalog number P1644-500ML |
| Dapi | Thermo Fisher | Catalog number D1306 |
| Growth-Factor reduced Matrigel | Corning | Catalog number 356230 |
| Type 1 collagen | Stem Cell Technologies | Catalog number 4902 |
| Rock Inhibitor | Stem Cell Technologies | Catalog number 72304 |
| IWR1-endo | cayman chemical | Catalog number 13659 |
| SB431542 | Tocris | Catalog number 1614 |
| Mitotracker Red | ThermoFisher | Catalog number M7512 |
| Recominant Midkine | EMD Millipore | Catalog number AG243 |
| Midkine inhibitor | Axon Medchem | Catalog number 2258 |
| Critical commercial assays | ||
| RNAscope Multiplex Fluorescence V2 Assay | ACD | Catalog number 323135 and 323180 |
| Chromium Single Cell 3’ GEM, Library and Gel Bead Kit V3 | 10x Genomics | Catalog number 1000075 |
| Chromium Chip B Single Cell Kit, 48 reactions | 10x Genomics | Catalog number 1000153/1000073 |
| Deposited data | ||
| Raw and analyzed prenatal human vascular data | This paper | GEO: GSE63473 |
| Experimental models: Cell lines | ||
| Human umbilical vein endothelial cells | ATCC | Catalog number CRL-1730 |
| NIH 3T3 | ATCC | CVCL 0594 |
| 1323-4 iPSC line | Gladstone Institute | Bruce Conklin lab |
| H28126 iPSC line | University of Chicago | Gilad |
| Oligonucleotides | ||
| RNAscope® Probe – Hs-ATP1A2-C2 | ACD | Catalog number 497451-C2 |
| RNAscope® Probe – Hs-TAGLN-C2 | ACD | Catalog number 498961-C2 |
| RNAscope® Probe – Hs-KCNJ8 | ACD | Catalog number 521891 |
| RNAscope® Probe – Hs-SERPING1 | ACD | Catalog number 869491 |
| RNAscope® Probe – Hs-PRDX3 | ACD | Catalog number 1062541-C1 |
| RNAscope® Probe – Hs-LUM-C2 | ACD | Catalog number 494761-C2 |
| RNAscope® Probe – Hs-ANGPT2-C2 | ACD | Catalog number 594511-C2 |
| RNAscope® Probe – Hs-FBLN5-C2 | ACD | Catalog number 593511-C2 |
| RNAscope® Probe – Hs-ADGRG6-C2 | ACD | Catalog number 480121-C2 |
| RNAscope® Probe – Hs-MYL9 | ACD | Catalog number 593741 |
| RNAscope® Probe – Hs-GJA4 | ACD | Catalog number 856221 |
| RNAscope® Probe – Hs-ANXA2 | ACD | Catalog number 855281 |
| RNAscope® Probe – Hs-ADM-C1 | ACD | Catalog number 582511 |
| Software and algorithms | ||
| Fiji | Schneider et al., 2012 | RRID:SCR_002285 |
| Imaris | Oxford Instruments | RRID:SCR_007370 |
| Prism | GraphPad Prism Version 8.4.3 | RRID:SCR_002798 |
| FlowJo | FlowJo Version 10.6.2 | RRID: SCR_008520 |
| RNAVelocity | Velocyto.py Version 0.17 and scvelo Version 0.2.4 | RRID:SCR_018167 |
| Adobe Illustrator | Adobe | RRID:SCR_010279 |
| R | The R foundation | RRID:SCR_001905 |
| RStudio | The R foundation | RRID:SCR_000432 |
| Cell Chat | CellChat Version 1.1.3 | RRID:SCR_021946 |
| Other | ||
| Cell browser for cells in the developing human brain | This paper | https://cells.ucsc.edu/?ds=vascular-dev |


