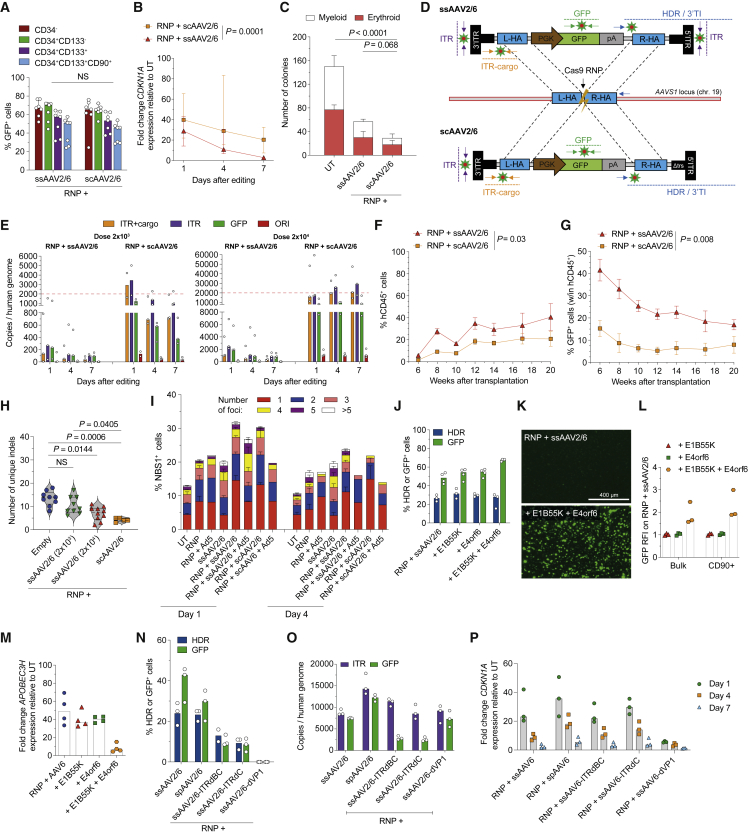
Figure 2.
AAV ITRs trigger p53 activation via MRN sensing
(A–C) Percentage of GFP+ cells within HSPC subpopulations (A, n = 7, Mann-Whitney test), fold change expression of CDKN1A over time relative to UT (B, n = 7, LME followed by post-hoc analysis; results are shown for the last time point), number of colonies grown from HSPCs (C, n = 9, Kruskal-Wallis test with Dunn’s multiple comparisons) after AAVS1 editing with the indicated AAVs. Median with 95% confidence interval (CI).
(D) Panel of digital droplet (dd) PCR probes tiling ss- and sc-AAV2/6 genomes (“ITR,” “ITR+cargo,” and “GFP”) and 3′ AAVS1 vector-genome junction (“3′ TI” or “3′ HDR”).
(E) Intracellular CG of the indicated AAV features retrieved over time from HSPCs edited with two doses of ss- or sc-AAV2/6 (n = 3). See STAR Methods for details. Median.
(F and G) Percentage of circulating hCD45+ (F) and GFP+ cells within the human graft (G) in mice transplanted with the outgrown progeny of starting-matched limiting cell doses of CB HSPCs edited as indicated (n = 5). Mean ± SEM, LME followed by post-hoc analysis; results are shown for the last time point.
(H) Number of unique indels retrieved by deep sequencing AAVS1 in human splenocytes from mice in Figures 1E and 1J and in (F) (n = 9, 9, 10, and 5). Median with quartiles. Kruskal-Wallis test with Dunn’s multiple comparisons.
(I) Quantification of NBS1+ cells and distribution of the percentage of foci bearing cells over time from HSPCs edited with the indicated treatments in the presence or absence of E1B55K and E4orf6 (Ad5; n = 11, 7, 1, 6, 6, 2, 1, and 3).
(J) Percentage of HDR-edited alleles and GFP+ cells in CB HSPCs edited in the presence or absence of E1B55K and/or E4orf6 (n = 3 for HDR and n = 4 for GFP). Median.
(K) Representative fluorescence microscopy images from (J).
(L) GFP relative fluorescence intensity (RFI) over the “RNP + ssAAV2/6” condition in bulk and primitive HSPCs from (J) (n = 3). Median.
(M) Fold change expression of APOBEC3H relative to UT 1 day after editing in experiments from (J) (n = 4). Median.
(N) Percentage of HDR-edited alleles and GFP+ cells in mPB HSPCs edited with different AAV variants as indicated (n = 3). Median.
(O) Intracellular CG of the indicated AAV features retrieved 1 day after treatment in HSPCs edited using different AAV variants (n = 3). See STAR Methods. Median.
(P) Fold change expression of CDKN1A over time relative to UT from experiments in (O) (n = 3). Median.
See also Figure S2.