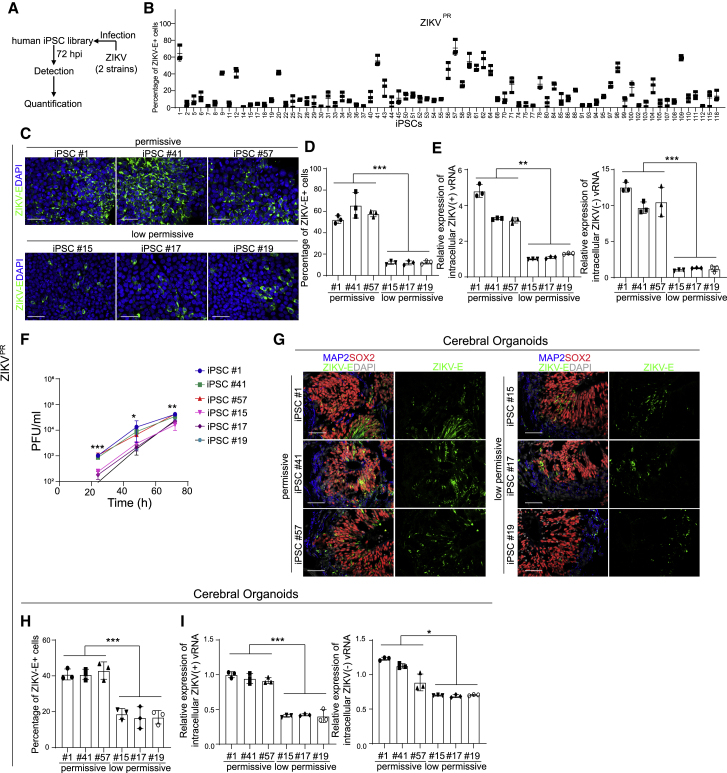
Figure 1.
An iPSC-array-based screen of ZIKV infection
(A) Scheme of the iPSC screening.
(B) The percentage of ZIKV-E positive cells of all iPSC lines upon ZIKVPR infection (ZIKVPR, Multiplicity of infection [MOI] = 1). Three biological replicates (each replicate includes one well of 96-well plate) were used to calculate the infectivity.
(C and D) Representative confocal images (C) and the quantification (D) of ZIKV-E staining of permissive cell lines iPSC #1, iPSC #41, and iPSC #57 and low-permissive cell lines iPSC #15, iPSC #17, and iPSC #19 at 72 hpi (ZIKVPR, MOI = 1). Scale bar, 50 μm.
(E) qRT-PCR analysis of (+) and (−) ZIKV vRNA strands of permissive cell lines iPSC #1, iPSC #41, and iPSC #57 and low-permissive cell lines iPSC #15, iPSC #17, and iPSC #19 at 72 hpi (ZIKVPR, MOI = 1). The data was normalized to actin beta (ACTB).
(F) Multiple step growth curve of ZIKV in the supernatant of permissive cell lines iPSC #1, iPSC #41, and iPSC #57 and low-permissive cell lines iPSC #15, iPSC #17, and iPSC #19 (ZIKVPR, MOI = 1).
(G and H) Representative confocal images (G) and the quantification (H) of ZIKV-E staining in cerebral organoids derived from permissive cell lines iPSC #1, iPSC #41, and iPSC #57 and low-permissive cell lines iPSC #15, iPSC #17, and iPSC #19 (ZIKVPR, 3 × 106 plaque-forming unit [PFU]/mL). Cerebral organoids were age-matched and collected at day 20, then infected with ZIKV for 24 h. After removal of virus-containing medium, organoids were maintained in organoid medium for an additional 3 days. Scale bar, 50 μm.
(I) qRT-PCR analysis of (+) and (−) ZIKV vRNA strands in cerebral organoids derived from permissive cell lines iPSC #1, iPSC #41, and iPSC #57 and low-permissive cell lines iPSC #15, iPSC #17, and iPSC #19 (ZIKVPR, 3 × 106 PFU/mL). Cerebral organoids were age-matched and collected at day 20, then infected with ZIKV for 24 h. After removal of virus-containing medium, organoids were maintained in organoid medium for an additional 3 days. The value was normalized to ACTB.
Data are representative of at least three independent experiments. Data are shown as mean ± SD. p values were calculated by unpaired two-tailed Student’s t test; ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001. See also Figure S1.