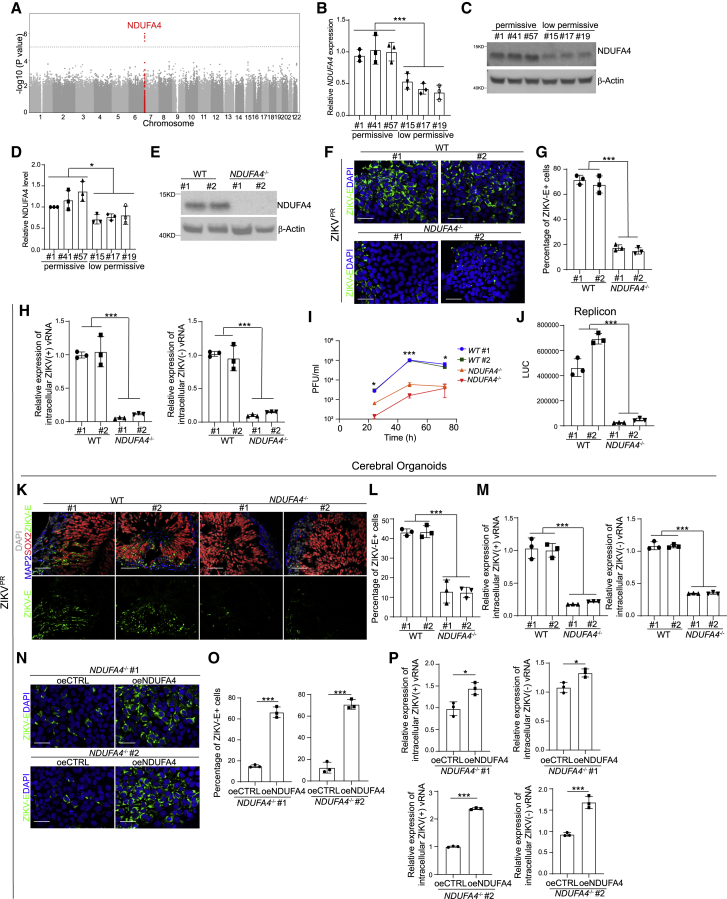
Figure 2.
Loss or reduction of NDUFA4 impairs ZIKV replication in vitro and in vivo
(A) Manhattan plot with highlighted risk locus.
(B) qRT-PCR analysis of NDUFA4 mRNA expression levels in permissive iPSC lines iPSC #1, iPSC #41, and iPSC #57 and low-permissive cell lines iPSC #15, iPSC #17, and iPSC #19. The value was normalized to ACTB.
(C and D) Western blotting analysis (C) and the quantification (D) of NDUFA4 protein expression levels in permissive iPSC lines iPSC #1, iPSC #41, and iPSC #57 and low-permissive cell lines iPSC #15, iPSC #17, and iPSC #19. β-Actin was used as a loading control.
(E) Western blotting analysis of NDUFA4 expression levels in WT or NDUFA4−/− hiPSCs. β-Actin was used as a loading control.
(F and G) Representative confocal images (F) and the quantification (G) of ZIKV-E staining in ZIKV-infected WT or NDUFA4−/− hiPSCs at 72 hpi (ZIKVPR, MOI = 1). Scale bar, 50 μm.
(H) qRT-PCR analysis of (+) and (−) ZIKV vRNA strands in ZIKV-infected WT or NDUFA4−/− hiPSCs at 72 hpi (ZIKVPR, MOI = 1). The value was normalized to ACTB.
(I) Multiple step growth curve of ZIKV virus in the supernatant of ZIKV-infected WT or NDUFA4−/− hiPSCs (ZIKVPR, MOI = 1).
(J) Luciferase activity of WT or NDUFA4−/− hiPSCs at 24 h after transfection with ZIKV replicon.
(K and L) Representative confocal images (K) and the quantification (L) of ZIKV-E staining in cerebral organoids derived from WT or NDUFA4−/− hiPSCs (ZIKVPR, 3 × 106 PFU/mL). Cerebral organoids were age-matched and collected at day 20, then infected with ZIKV for 24 h. After removal of virus-containing medium, organoids were maintained in organoid medium for an additional 3 days. Scale bar, 50 μm.
(M) qRT-PCR analysis of (+) and (−) ZIKV vRNA strands in cerebral organoids derived from WT or NDUFA4−/− hiPSCs at 72 hpi (ZIKVPR, 3 × 106 PFU/mL). Cerebral organoids were age-matched and collected at day 20, then infected with ZIKV for 24 h. After removal of virus-containing medium, organoids were maintained in organoid medium for an additional 3 days. The value was normalized to ACTB.
(N and O) Representative images (N) and the quantification (O) of ZIKV-E staining in ZIKV-infected NDUFA4−/−-oeCTRL and NDUFA4−/−-oeNDUFA4 hiPSCs at 72 hpi (ZIKVPR, MOI = 1). Scale bar, 50 μm.
(P) qRT-PCR analysis of (+) and (−) ZIKV vRNA strands in ZIKV-infected NDUFA4−/−-oeCTRL and NDUFA4−/−-oeNDUFA4 hiPSCs at 72 hpi (ZIKVPR, MOI = 1). The value was normalized to ACTB.
Data are representative of at least three independent experiments. Data are shown as mean ± SD. For comparison of permissive and low-permissive lines, p values were calculated by unpaired two-tailed Student’s t test. For comparison of WT and KO lines, p values were calculated by two-way ANOVA analysis. For comparison of control and overexpression groups, p values were calculated by unpaired two-tailed Student’s t test; ∗p < 0.05, ∗∗∗p < 0.001. See also Figure S2.