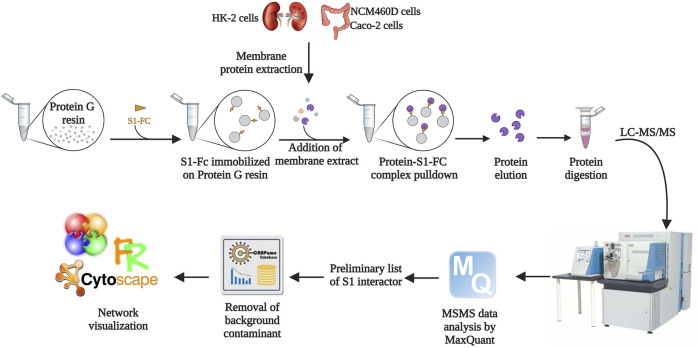
FIGURE 1.
Workflow of the AP-MS approach followed in the present study. The solid support was functionalized with a recombinant form of S1. Membrane protein extracts derived from HK-2, NCM460D, and Caco-2 cell lines were incubated with the resin, and the retained protein complexes were eluted in denaturing conditions. The samples were subjected to a shotgun hydrolysis protocol using trypsin as the proteolytic enzyme. The peptide mixtures were analyzed by nano LC-MS/MS and proteins were identified by the software MaxQuant. The preliminary lists obtained discarding the proteins identified in the negative controls were further filtered with the contaminant database CRAPome. The final lists were analyzed by bioinformatics tools such as Cytoscape.