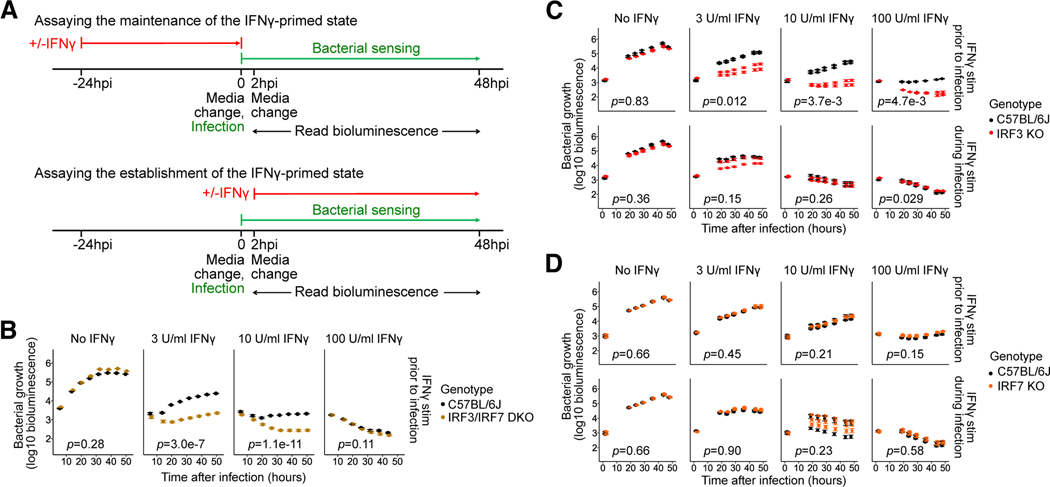
Fig. 1: Contribution of IRF3 and IRF7 to the maintenance and establishment of the IFNγ-activated state in BMMs.
(A) Design of the L. pneumophila infection assay to assess the effects of genetic perturbations on the maintenance (top) or establishment (bottom) of the IFNγ-activated state.
(B-D) BMMs from B6 and (B) Irf3−/−/Bcl2L12−/−/Irf7−/− (labeled IRF3/IRF7 DKO), (C) Irf3−/−/Bcl2L12−/− (labeled IRF3 KO), and (D) Irf7−/− (labeled IRF7 KO) mice were stimulated with IFNγ at concentrations of 0, 3, 10, or 100U/ml either before infection (i.e., ‘primed’) (B, C-D top panels) or at 2hpi (C-D bottom panels), then infected with L. pneumophila. Bacterial bioluminescence was measured over two days of infection. In (B, C, and D), data points show represent biological replicates (mice) (one, two, and three, respectively) and error bars represent the standard error of the mean across technical replicates (wells) (four, five, and five, respectively). p-values indicate the significance of the effect of genotype on bacterial growth over time under the conditions in each sub-panel, based on a linear mixed-effect model.