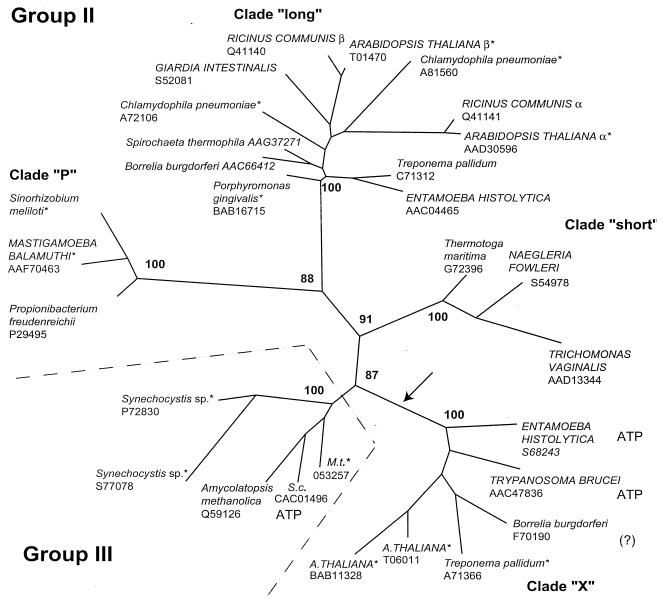
FIG. 1.
Unrooted phylogenetic tree of group II and III PPi-PFKs based on 167 shared amino-terminal residues (between residues 2 and 204 in the P. freudenreichii sequence). Taxon selection extended over the complete diversity present in available databases. The arrow denotes a proposed gene duplication. Close paralogs among chlamydiae, actinomycetales, and plants were omitted for clarity. Database accession numbers follow the species names. For the S. meliloti sequence, see http://sequence.toulouse.inra.fr/meliloti.html. Eukaryotic species names are in capital letters, and eubacterial species names are in lowercase letters. A., Arabidopsis; M.t., Mycobacterium tuberculosis; S.c., Streptomyces coelicolor. PPi specificity was established biochemically for unmarked species; asterisks mark species not explored biochemically, “ATP” indicates enzymes with ATP specificity, and the question mark indicates a species for which the heterologously expressed enzyme showed no PFK activity with either PPi or ATP (9). For the long clade plant enzymes, α is the regulatory subunit and β is the catalytic one. The maximum-likelihood method with local rearrangements was utilized. Numbers at the nodes give RELL bootstrap values (see text).