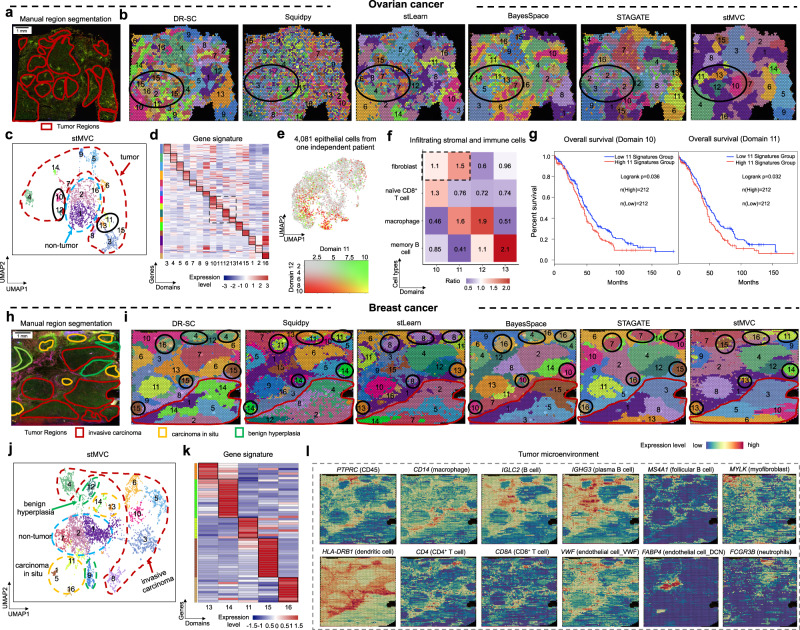
Fig. 3. stMVC enables the detection of cell-states distributed in different spatial domains on ovarian and breast cancers.
a Immunofluorescent staining of the tissue section and 17 manually segmented tumor regions. The intensity of DAPI, cytokeratin, and CD45 is shown in blue, green, and yellow. b Spatial clustering by DR-SC, Squidpy, stLearn, BayesSpace, STAGATE, and stMVC. c UMAP visualization of latent features by stMVC. Each domain is outlined by the region it belongs to, i.e., tumor or non-tumor. d Heatmap of average gene expression of signature genes from 16 domains by stMVC. Rows and columns indicate genes and domains. e UMAP plot of average expression of signature genes for domains 11 and 12 in scRNA-seq data of 4081 epithelial cells from one independent ovarian cancer. f Enrichment of infiltrating stromal and immune cells in each domain compared to the total distribution of those in four domains. The ratio is calculated by chi-square test85. The larger the ratio, the more cells are enriched in the domain. g Total survival rate of patients with the high ( median value) or low (low median value) expression level of 11 signature genes for domains 10 and 11 in gene expression data of ovarian cancer from TCGA by GEPIA286 (Supplementary Table 1). h Immunofluorescent staining of the tissue section and 15 manually segmented tumor regions, where those regions are outlined by their histological annotations: invasive carcinoma (red), carcinoma in situ (yellow), and benign hyperplasia (green). The intensity of DAPI, fiducial frame, and anti-CD3 is shown in green, blue, and yellow. i Spatial clustering by DR-SC, Squidpy, stLearn, BayesSpace, STAGATE, and stMVC. j UMAP visualization of latent features by stMVC. Domains are outlined by their histological annotations. k Heatmap of gene expression of signature genes from five domains enriched in the carcinoma in situ region by stMVC. Rows and columns indicate genes and domains. l Spatial expression of classical marker genes for stromal and immune cells, including PTPRC, CD14, IGCL2, IGHG3, MS4A1, MYLK, HLA-DRB1, CD4, CD8A, ENG, FABP4, and FCGR3B for data denoised by stMVC. Source data are provided as a Source Data file.