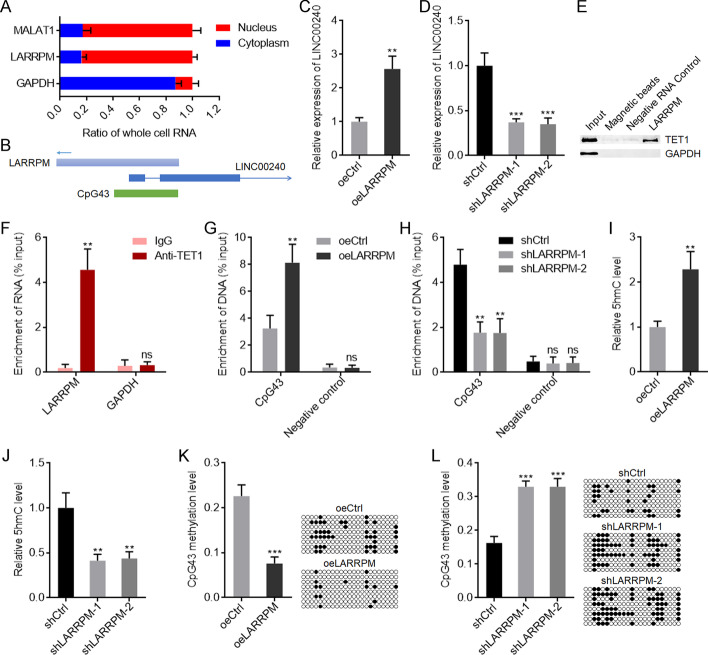
Fig. 4.
LARRPM upregulated LINC00240 expression via inducing DNA demethylation. a Subcellular localization of LARRPM in A549 cells was detected using subcellular fractionation followed by qRT-PCR. MALAT1 and GAPDH were used as nuclear and cytoplasmic controls, respectively. b The schematic diagram of the genomic localization of LARRPM and LINC00240. c, d LINC00240 expression in A549 cells with LARRPM overexpression (c) and HCC827 cells with LARRPM depletion (d) was detected by qRT-PCR. e RNA–protein pull-down assays followed by western blot were performed to detect the proteins bound by LARRPM. f RNA immunoprecipitation (RIP) assays followed by qRT-PCR were performed to detect the RNAs bound by TET1. g, h Chromatin immunoprecipitation (ChIP) assays followed by qPCR were performed in A549 cells with LARRPM overexpression (g) and HCC827 cells with LARRPM depletion (h) to detect the DNAs bound by TET1. i, j 5-Hydroxymethylcyctosine (5hmC) levels of CpG43 from A549 cells with LARRPM overexpression (i) and HCC827 cells with LARRPM depletion (j) were measured using the EpiMark 5-hmC Analysis Kit. k, l DNA methylation levels of CpG43 from A549 cells with LARRPM overexpression (k) and HCC827 cells with LARRPM depletion (l) were measured using bisulfate DNA sequencing. Results are shown as the mean ± SD based on three independent experiments. Asterisks indicate a significant difference at **P < 0.01, ***P < 0.001; ns, not significant, by Student’s t-test (c, f, g, i, k) or one-way ANOVA followed by Dunnett's multiple comparisons test (d, h, j, l). GAPDH Glyceraldehyde 3-phosphate dehydrogenase, LINC00240 a long LUAD-related non-coding RNA, MALATI a long non-coding RNA, TET1 a DNA demethylase