Figure 1.
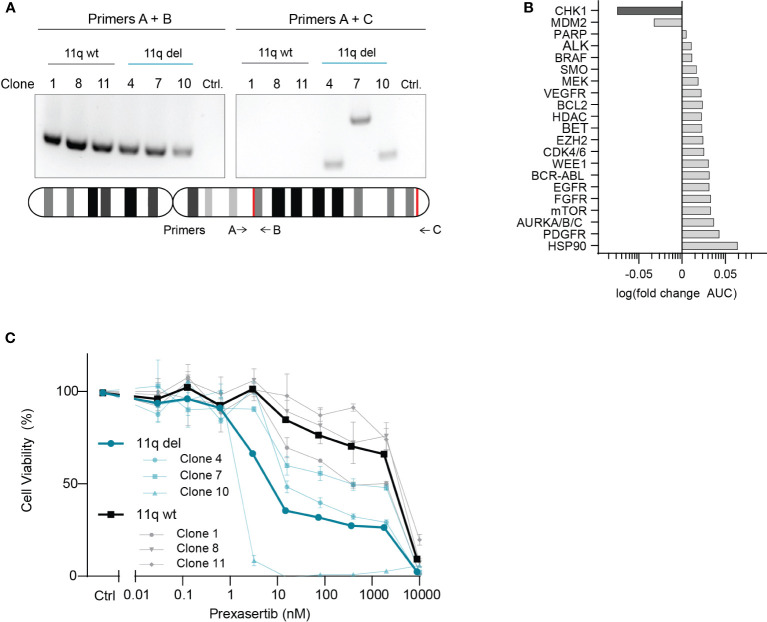
CHK1 identified as a therapeutic target in 11q deleted NB. (A) Agarose gel electrophoresis of PCR products from 11q wild type (black) and 11q deleted (blue) SKNSH clones using primer set A + B (left) to amplify the wild type copy of 11q, and primer set A + C (right) which only yields a product if 11q deletion was induced. Shown below is an ideogram of chromosome 11q with the approximate location of primer targets and the gRNA recognition sites for CRISPR-Cas9 directed 11q deletion (red). (B) Median log fold change in area under the curve (AUC) for 11q deleted and wild type SKNSH clones following 72-hour incubation with compounds targeting the proteins listed on the y-axis. (C) Dose-response curves for three SKNSH clones with chromosome 11q deletion (Clone 4, 7, 10; blue) and three SKNSH clones with a normal chromosome 11q locus (Clone 1, 8, 11; grey). Average dose-response curves for each phenotype are indicated in bold. All curves represent the average of replicates (n=2), and error bars indicate the standard error of the mean (SEM).