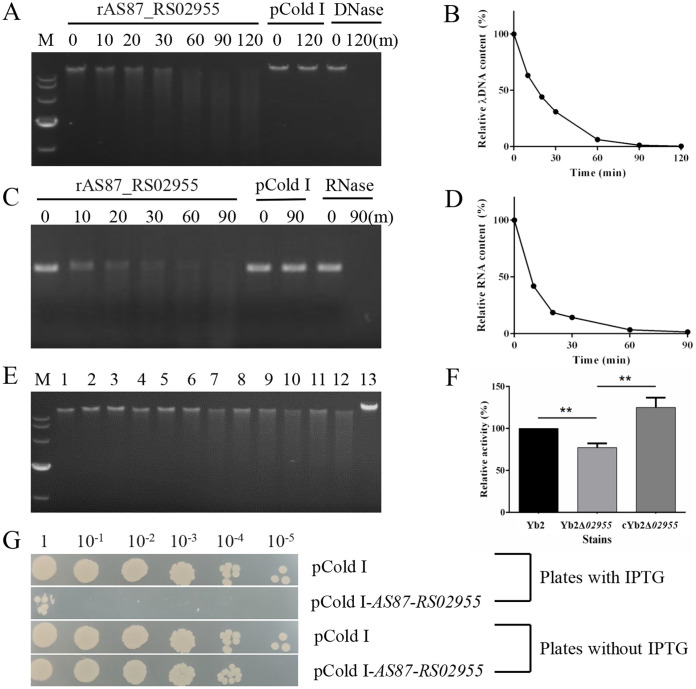
FIG 5.
Enzymatic activity assays of R. anatipestifer recombinant AS87_RS02955 (rAS87_RS02955). (A) Enzymatic activity of rAS87_RS02955 in cleaving λ DNA. (B) λ DNA cleavage curve. (C) Enzymatic activity of rAS87_RS02955 in cleaving MS2 RNA. (D) MS2 RNA cleavage curve. Purified rAS87_RS02955 was incubated with substrates at 37°C for 0, 10, 20, 30, 60, 90, and 120 min (except 120 min for RNA) in the presence of Mg2+. Purified pCold I was used as a negative control. DNase and RNase were used as positive controls. Cleavage curves were generated using Image J software. (E) Enzymatic activity of R. anatipestifer secretory proteins. Lane M, DL10,000 DNA marker; lanes 1 to 3, 1 h reactions; lanes 4 to 6, 3 h reactions; lanes 7 to 9, 6 h reactions; lanes 10 to 12, 9 h reactions. Lanes 1, 4, 7, and 10, secretory proteins from Yb2; lanes 2, 5, 8, and 11, secretory proteins from Yb2Δ02955; lanes 3, 6, 9, and 12, secretory proteins from cYb2Δ02955; lane 13, no protein reaction for 9 h at 37°C. (F) Relative enzymatic activity of secretory proteins after incubation for 9 h. Yb2 activity toward λ DNA was 100%: Yb2Δ02955 showed 70% relative activity compared with Yb2. Complementation with AS87_RS02955 recovered enzymatic activity. Error bars represent the standard deviation of data from three independent experiments (**, P < 0.01). (G) AS87_RS02955 activity in E. coli BL21(DE3) cells. Cells were transformed with pCold I-AS87_RS02955, grown to logarithmic phase, washed twice in PBS, adjusted to OD600 0.8, 10-fold serially diluted, and 2 μL spotted onto LB agar plates with/without IPTG for overnight culture at 37°C. Cells transformed with the pCold I vector were used as a control strain. “1” means 2 μL bacteria at OD600 = 0.8; 10−1, 10−2, 10−3, 10−4, and 10−5 indicated 10-fold series dilution of the bacteria at OD600 = 0.8.