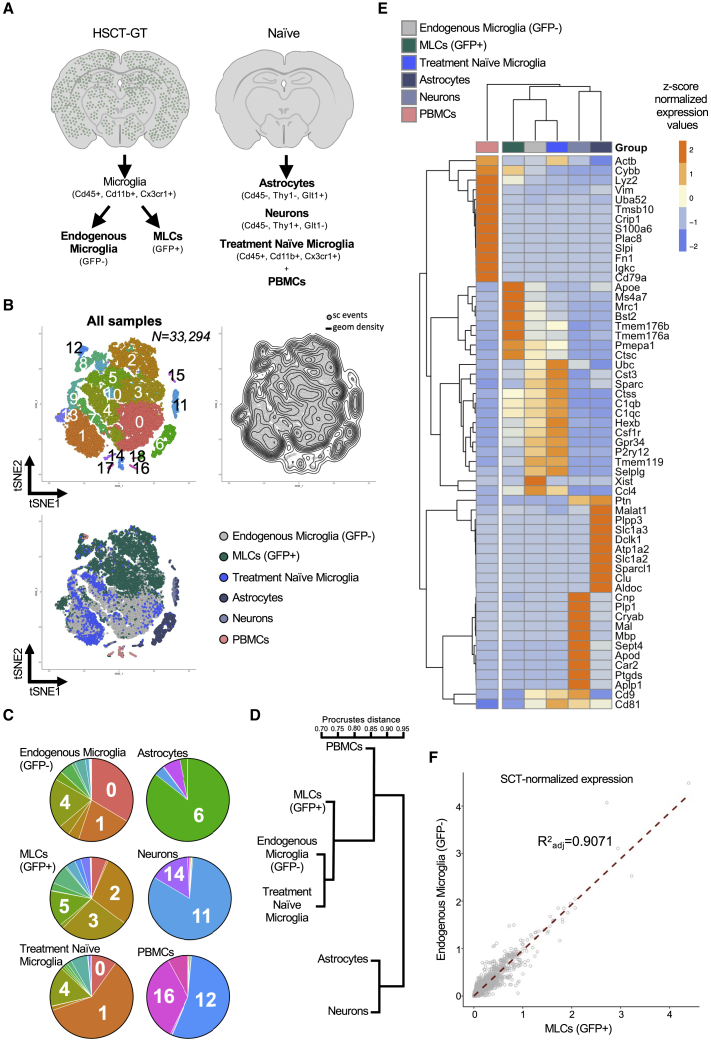
Figure 3.
Single-cell comparison of MLCs, endogenous microglia, PBMCs, neurons, and astrocytes
(A) MLCs and endogenous microglia were isolated via enzymatic digestion, FACS purified, and then single-cell sequenced from i.v.- and i.c.v.-dosed male and female animals. PBMCs, neurons, astrocytes, and microglia were similarly isolated from a treatment-naive animal. (B) Global tSNE plot generated from a total of 33,294 cells (n = 5 mice; 12 samples). The plot on the left shows the Louvain clustering (resolution 0.4), while the plot on the right shows the density of single-cell events on the map. (C) Pie charts showing the cluster breakdown by sample, with labels showing the top represented clusters for each sample type. Seven MLC-enriched clusters (2–5, 8, 10, 12, 13, 15), 5 microglia-enriched clusters (0, 1, 4, 7, 9), 1 neuron cluster (11), 1 astrocyte cluster (6), and 4 PBMC clusters (14, 16–18) were identified. (D) Unsupervised clustering based on the global expression profile of each sample type. (E) Heatmap showing top 10 differentially expressed genes for each sample type. Dendrogram at the top shows samples clustering based on expression profiles of these genes. (F) Correlation analysis of endogenous microglia versus MLCs based on normalized single-cell gene-expression data.