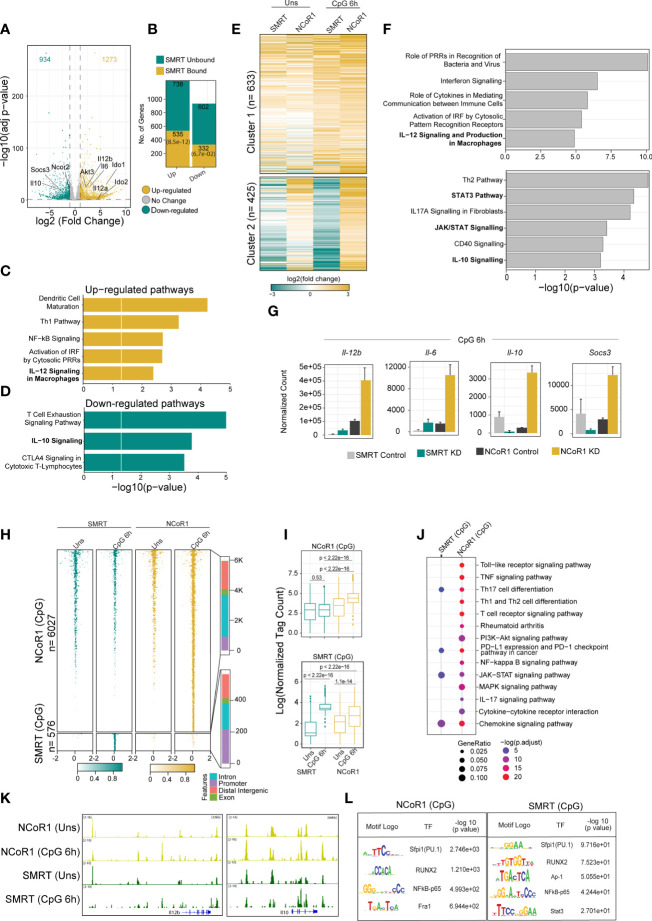
Figure 6.
Integrative genomics analysis (RNA-seq and ChIP-seq) identified the differential role of SMRT and NCoR1 in regulation of immune response in cDC1. (A) Volcano plot showing the differentially expressed genes (DEGs) in SMRT KD cDC1 DCs as compared to control cells after 6h CpG stimulation. 1273 and 934 genes upregulated and downregulated respectively upon SMRT depletion (n=5). (B) Bar plot showing the total number of SMRT bound and unbound DEGs in unstimulated and 6h CpG stimulated SMRT KD cDC1 DCs. p-value showing significance of overlap between SMRT bound genes and DEGs. (C) Bar plot depicting enriched canonical pathways for the list of SMRT bound upregulated DEGs in 6h CpG activated SMRT KD cDC1 DCs as compared to control cells using Ingenuity pathway analysis (IPA). (D) Bar plot depicting enriched canonical pathways for the list of significantly SMRT bound down-regulated DEGs in 6h CpG activated SMRT KD cDC1 DCs as compared to control cells using Ingenuity pathway analysis (IPA). (E) Heatmap showing two clusters obtained from K-means clustering of log2 fold change of DEGs upon SMRT and NCoR1 KD compared to control cells in unstimulated and 6h CpG stimulation condition. (F) Bar plot showing enriched canonical pathways from IPA for respective clusters shown in Fig 6E. (G) Bar plot with standard deviation showing normalized count from DESeq2 of selected genes from enriched pathway for cluster-1 (Il12b and Il6) and cluster2 (Il10 and Socs3). (H) Tornado plot showing ChIP-seq signal ( ± 2kb to peak center) of differential NCoR1 and SMRT binding sites (NCoR1 CpG and SMRT CpG) in unstimulated and 6h CpG stimulation (1st panel). Bar plot showing the distribution of differential genomic regions based on distance relative to TSS (2nd panel). (I) Box plot showing normalized tag count in differential NCoR1 CpG and SMRT CpG cluster. Boxes encompass the 25th to 75th percentile of normalized tag count. Whiskers extend to the 10th and 90th percentiles. Mean difference significance was calculated using the Wilcoxon test. (J) Dot plot showing the significantly enriched KEGG terms for genes associated with NCoR1 and SMRT CpG binding clusters shown in Fig 6I. KEGG term enrichment analysis was performed using cluterProfiler R package. (K) IGV snapshot showing NCoR1 and SMRT binding on Il12b and Il10 gene loci. (L). Table showing transcription factor motifs that were significantly enriched (P-value< 1e-10) in differential NCoR1 CpG and SMRT CpG clusters.