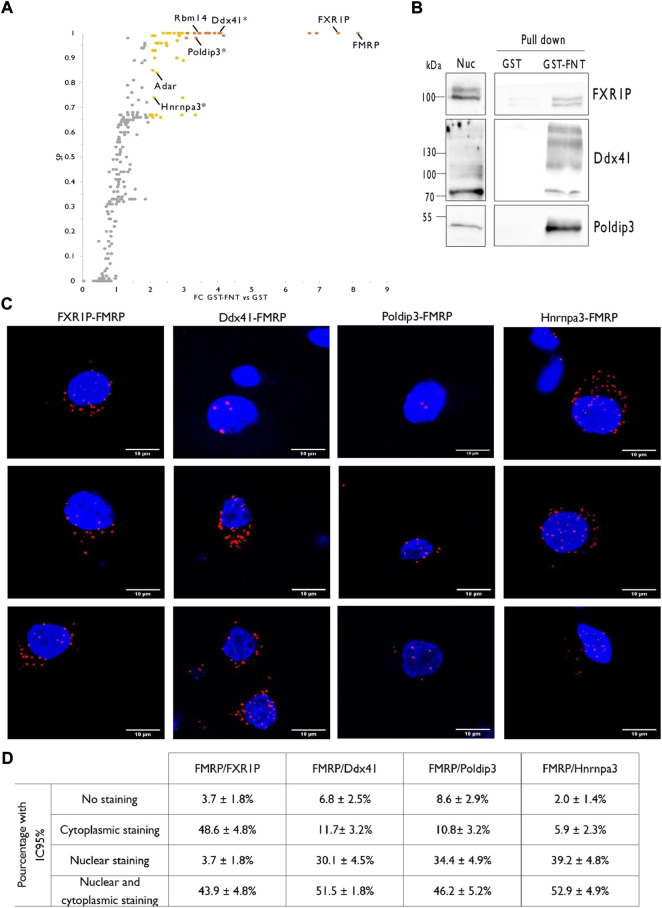
FIGURE 3.
Identification of rat forebrain nuclear FMRP-interacting proteins. (A). Volcano plot highlighting proteins differentially co-purified in triplicate pull-down experiments using the GST-FNT recombinant protein versus the GST alone. Statistically significant differences were assessed using the Saint-Express tool. Unenriched proteins are displayed in gray, while enriched proteins presenting a FC > 3.00 and SP > 0.7 are displayed in orange, and were classified as “high-confidence” interactor. Yellow dots corresponding to all other significant proteins with FC > 2.0 and SP > 0.5, and were referred as “medium-confidence” interactor. Know and novel (*) FMRP interacting proteins are noted in the scatter plot. (B). Immunoblot analysis of identified FMRP-interacting proteins. Pull down experiments were conducted with GST-FNT and GST (negative control) immobilized on Glutathione Sepharose with nuclear fractions. Protein retained on the beads were resolved by SDS-PAGE and processed for western blotting using antibodies against FXR1P, Poldip3 or Ddx41. (C). Representative confocal images of the interaction between endogenous FXR1P, Ddx41, Poldip3 or Hnrnpa3, and FMRP detected by Proximity Ligation Assay (PLA) in rat primary hippocampal neuron cultures. Neuronal cells were identified upon their MAP2 labelling (not shown). (D). Percentage of neurons presenting no PLA dots (No staining), PLA dots exclusively in the cytoplasm (Cytoplasmic staining), exclusively in the nucleus (Nuclear staining) or in both the cytoplasm, and the nucleus (Cytoplasmic and nuclear staining) for the indicated interactions. The percentages with the 95% Confidence Interval were calculated from 107 (FMRP/FXR1P), 103 (FMRP/Ddx41), 93 (FMRP/Poldip3), and 103 (FMRP/Hnrnpa3) neurons in culture processed from three independent experiments.