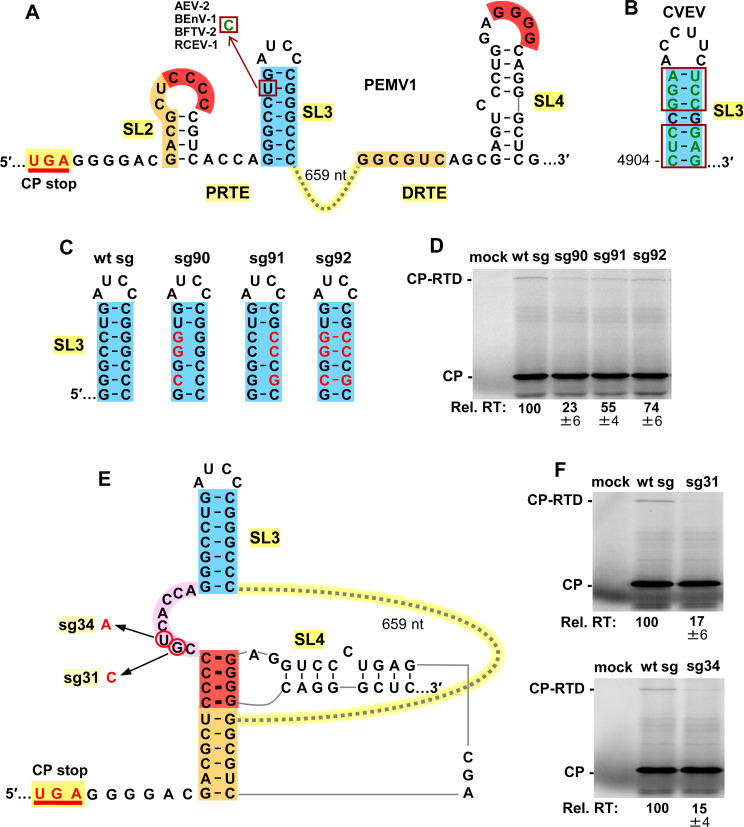
Fig 5. Assessing the local SL3 (blue) in the PRTE.
(A) RNA secondary structures of PRTE and DRTE in PEMV1. A nucleotide mono-variation (U to C) in the SL3 of four enamoviruses that maintains base pairing is shown (boxed). Alfalfa enamovirus-2 (AEV-2, KY985463.1), bean enamovirus-1 (BEnV-1, MZ361924), birdsfoot trefoil virus-2 (BFTV-2, NC_048296) and red clover enamovirus-1 (RCEV-1, MN412742). (B) SL3 of citrus vein enation virus (CVEV, NC_021564). Covariations in the SL3 stem that maintain pairing are boxed. (C) Compensatory mutations introduced into SL3, with substitutions depicted in red. (D) Results of in vitro translation reactions for the sg mRNAs shown in panel C. Averaged Rel. RT levels (±SE) calculated from three independent trials are shown below each lane. (E) Proposed RNA secondary structure when the red and orange long-distance interactions between the PRTE and DRTE occur. The linker sequence between red and blue helices is highlighted in pink, with corresponding substitutions in mutant sg mRNAs circled and indicated in red. (F) Results of in vitro translation reactions for mutant sg mRNAs shown in panel E. Averaged Rel. RT levels (±SE) calculated from three independent trials are shown below each lane.