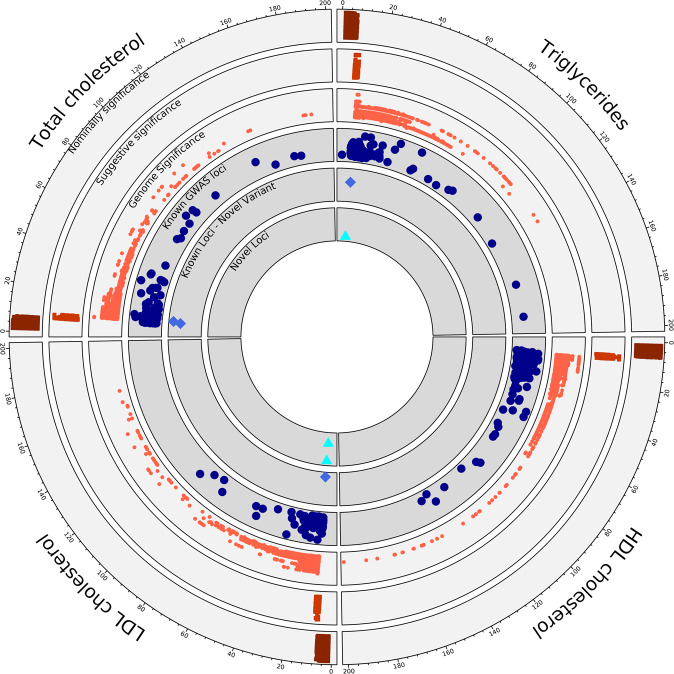
Fig. 2. Summary of single variant genome-wide association.
Representation of the single variant GWAS results from TOPMed Freeze 8 whole genome sequenced data of 66,329 samples. Each quarter represents a different lipid phenotype, and dots extending in clock-wise fashion represent variants with increasing evidence of association as noted by −log10(p-value), which was truncated at 200. The outer three circles show the GWAS data from TOPMed freeze8 where variants binned to nominally significant (p-value 0.05–5 × 10−07), suggestive significant (p-value 5 × 10−07–5 × 10−09) and genome wide significant (p-value < 5 × 10−09). The inner three circles compare our TOPMed results with known significantly associated lipid loci and variants from the MVP summary statistics and GWAS catalog to the identified novel variants and loci that are genome-wide significant from the current study, respectively. The figure represents the outputs from two-sided genetic association testing preformed using SAIGE-QT model, where the model was adjusted for all the covariates; see Methods. TOPMed Trans-Omics for Precision Medicine, GWAS genome wide association study, MVP million veteran program.