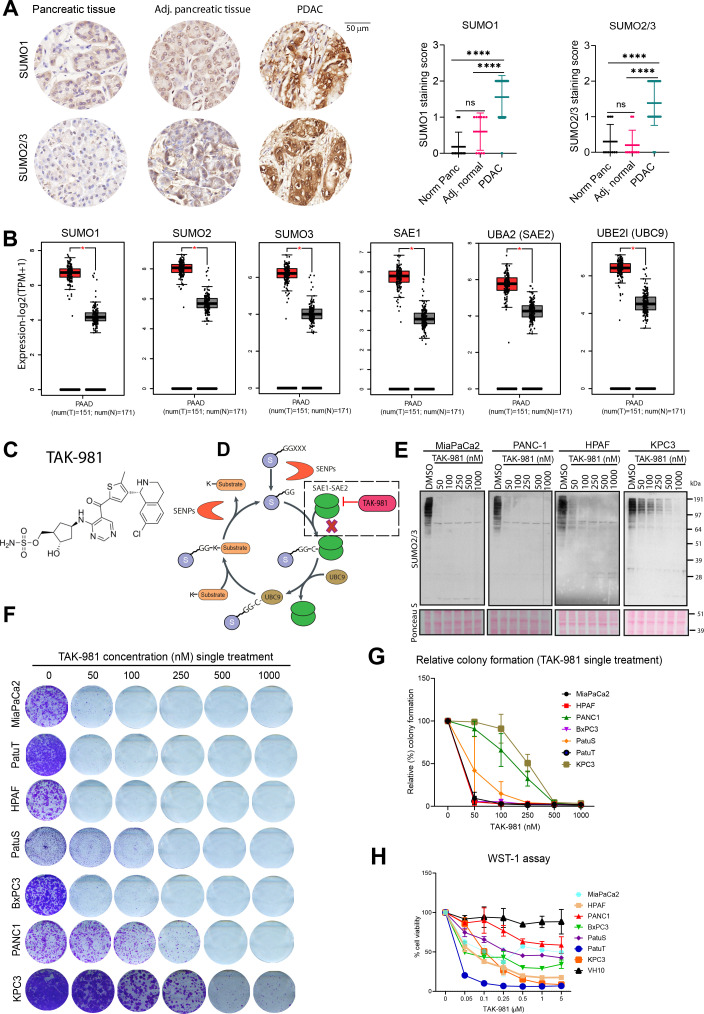
Figure 1.
SUMO pathway components are overexpressed in PDAC and required for tumour cell proliferation. (A) Immunohistochemical analysis of SUMO1 and SUMO2/3 in PDAC, adjacent normal and normal pancreatic tissue from tissue microarray of PDAC. Dot plots represent the quantification of SUMO1 and SUMO2/3 immunodetection. P values were calculated using one-way ANOVA with Tukey’s multiple comparison test. ****p<0.0001; n.s. indicates non-significant p value. (B) SUMO pathway genes mRNA expression in PDAC (red) and normal pancreas (grey) based on the TCGA and GTEx data analysed by GEPIA2. *p<0.05. (C) chemical structure of TAK-981. (D) SUMOylation cycle and the inhibition of SUMO-activating enzyme (SAE) by TAK-981. (E) Western blot analysis of SUMO2/3 levels of the human pancreatic cancer cell lines MiaPaCa2, PANC1, HPAF and mouse pancreatic cancer cell line KPC3, treated for 4 hours with the indicated concentrations of TAK-981. Ponceau S staining was used as loading control (n=2). (F) Multiple human pancreatic cancer cell lines MiaPaCa2, PANC1, HPAF, BxPC3, PatuS, PatuT and mouse pancreatic cell line KPC3 were treated with the indicated concentrations of TAK-981 for 4 days and colony formation was determined by crystal violet staining. (G) Line graphs represent the absorbance of solubilised crystal violet as mean with standard deviation (n=3). (H) Cell viability of human pancreatic cells and foreskin fibroblast primary cells was measured with the WST-1 assay. Cells were treated with the indicated concentrations of TAK-981 for 4 days. Relative cell viability is represented as mean with SD. The experiment was carried out twice in triplicate (n=6). ANOVA, analysis of variance; GTEx, genotype-tissue expression; PDAC, pancreatic ductal adenocarcinoma; SUMO, small ubiquitin like modifiers; TCGA, The Cancer Genome Atlas.