Fig. 3 |. Novel methods accelerate RNA sensor selection.
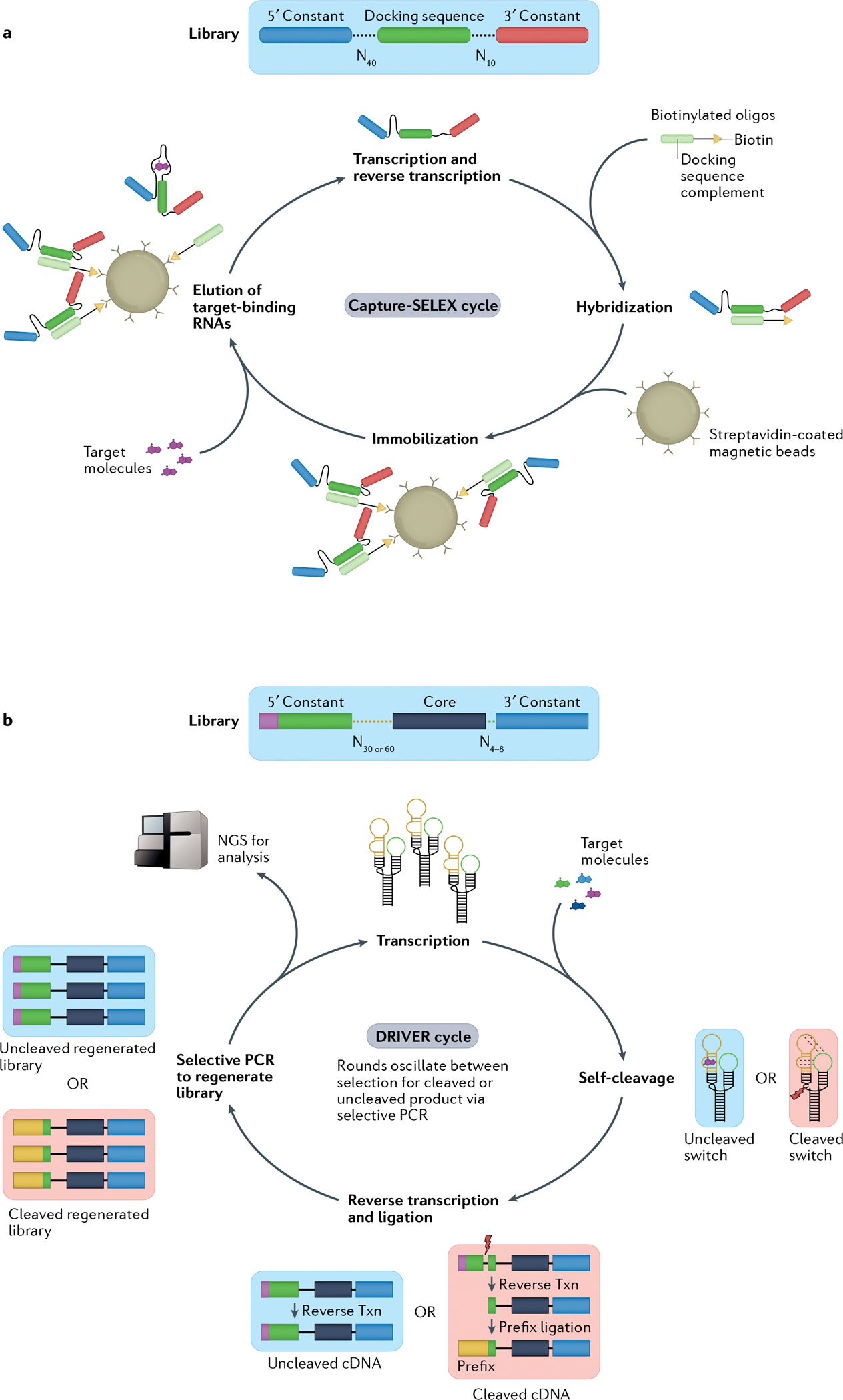
Overview of two methods for selection of RNA aptamers or sensors. Both methods begin with a diverse DNA library consisting of 1012–1015 distinct members. a | In the Capture-SELEX (systematic evolution of ligands by exponential enrichment) method23,84,85, initial sequences consist of a 5′ and 3′ constant region flanking two randomized regions of 10 or 40 nucleotides with a constant docking sequence in the centre. The transcribed library is then hybridized to a biotinylated oligonucleotide via base pairing of the docking sequence. The complexes are immobilized on streptavidin-coated magnetic beads and mixed with the target molecules. RNA sequences that bind the target molecules and undock from the biotinylated oligonucleotides are subsequently eluted from the beads, creating the starting library for the next selection cycle. b | In the DRIVER (de novo rapid in vitro evolution of RNA biosensors) method81, libraries are designed based on a modified hammerhead ribozyme (HHRz) from satellite RNA of tobacco ringspot virus (sTRSV) — a small, naturally occurring self-cleaving ribozyme — with the two loops replaced by either a randomized 30–60mer or a randomized 4–8mer. Sequences are mixed with target molecules. Desired RNA biosensor sequences cleave in the absence of the target molecule but bind to the target molecule when present, preventing cleavage. In iterative cycles of negative and positive selection, the RNA sequences are reverse transcribed (txn) and the resultant cDNA of cleaved sequences has a 5′ primer ligated that allows for selective PCR amplification of the sequences corresponding to either the cleaved or uncleaved RNA. The amplified DNA library then serves as the starting library for a new round of DRIVER. Periodically, these libraries can be analysed following quantification by next-generation sequencing (NGS). Part a reprinted with permission from REF85, Elsevier.
