Fig. 4 |. Emerging mechanisms of RNA processing and cell control for novel RNA devices.
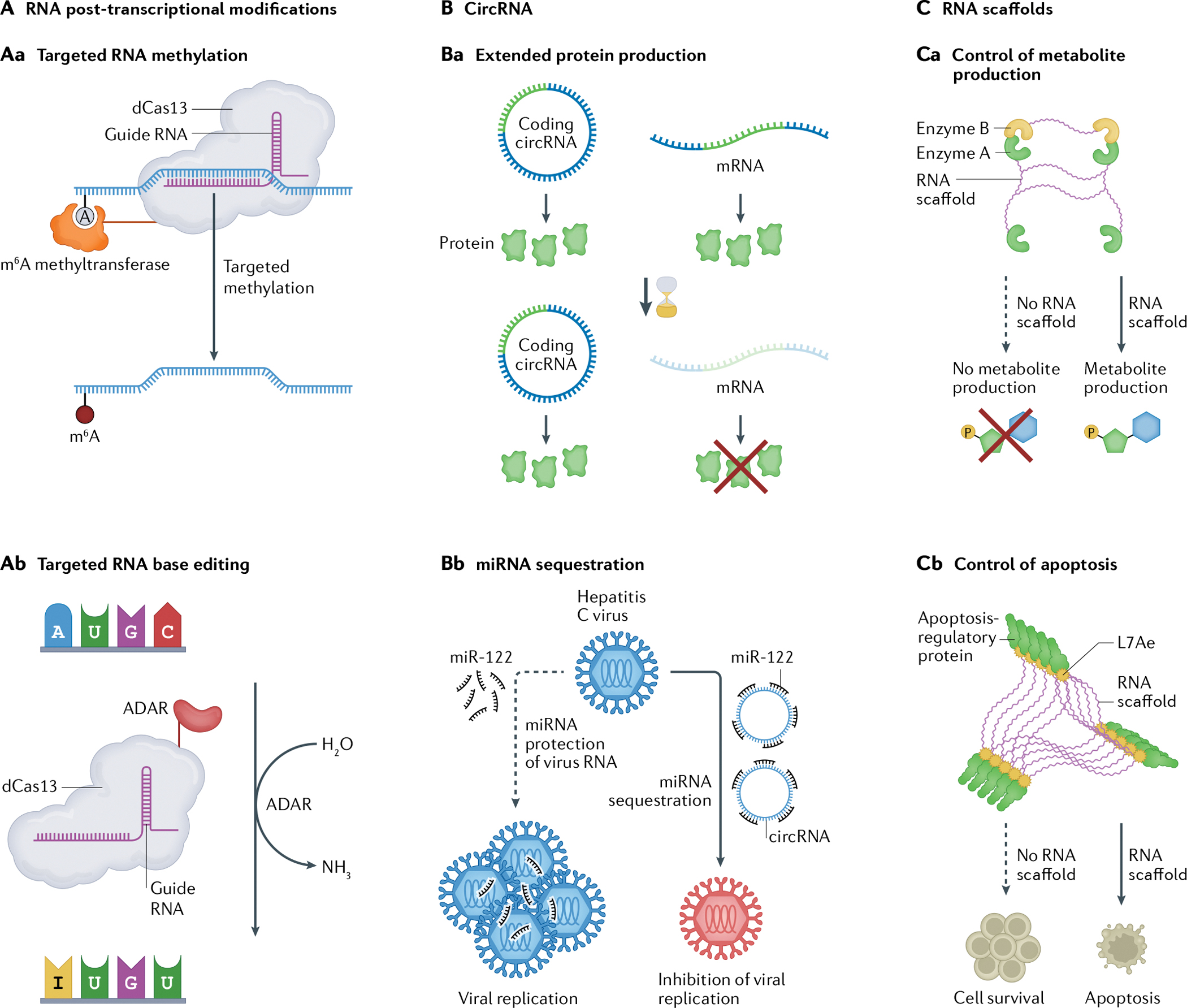
Recent findings have revealed new roles and mechanisms for RNA beyond control of translation through the central dogma, including targeted RNA base editing and post-transcriptional modifications (part A), circular RNA (circRNA) mechanisms (part B) and novel uses of RNA scaffolds (part C). Future RNA devices can incorporate these new mechanisms. Aa | Post-transcriptional modifications are involved in RNA regulation. N6-Methy[adenosine (m6A) is a post-transcriptional modification affecting mRNA stability and translation. Methyltransferases can be fused to dCas13 (a catalytically inactive CRISPR-associated nuclease) directed by a guide RNA to catalyse the conversion of adenosines to m6A, allowing targeted artificial methylation of RNA which could be further utilized99. Ab | RNA base editors use naturally occurring and evolved enzymes to modify RNA nucleobases for post-transcriptional gene editing. Current editors utilize deaminases to convert adenosine and cytosine to inosine and uracil, respectively103,104,106,152. Ba | circRNA expression vectors express proteins for longer than equivalent mRNA sequences107. Bb | Synthetic circRNAs can be designed as microRNA (miRNA) sponges to quench specific miRNAs and inhibit miRNA-dependent viral replication120. Ca | RNA can be repurposed as a scaffold to co-localize enzymes and increase local enzyme concentrations for control of metabolite production125. Cb | RNA scaffolds can also be constructed to induce proximity oligomerization of Caspase 8 within the caspase pathway for apoptosis126. ADAR, adenosine deaminase acting on RNA. Part Aa is adapted from REF99, Springer Nature Limited. Part Cb is adapted from REF126, CC-BY 4.0 (https://creativecommons.org/licenses/by/4.0/).
