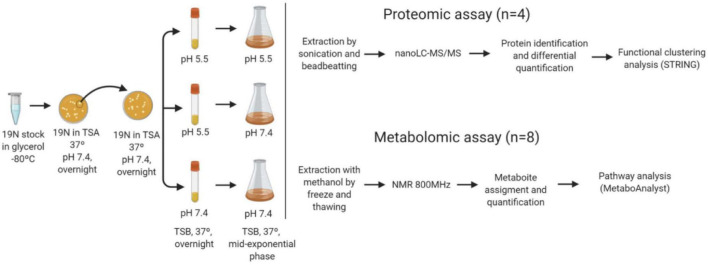
FIGURE 1.
Scheme of the workflow followed. Cells from –80°C glycerol stocks were streaked in Tryptic Soy agar media (TSA) and a single colony was picked to inoculate a new TSA plate. From the plate a single colony was collected to prepare a pre-inoculum in TS broth medium with adjusted pH (pH 5.5 and 7.4) and incubated overnight at 37°C and 225 rpm. The inoculum at pH 5.5 was used for the cultures grown at 5.5 (N55) and 7.4 (N57), and the inoculum at pH 7.4 for the culture at the same pH (N77). The cells were harvested at mid-exponential phase. For proteomics the proteins were extracted from 4 biological replicates/experimental condition and analysed by nanoLC-MS/MS. Identified proteins (MASCOT) were quantified by spectral counts. Differential abundant proteins between experimental conditions were submitted to functional clustering analysis (STRINGdb). Metabolites from eight replicates/experimental condition were extracted, after metabolic quenching and biomass normalisation. Metabolites were assigned in 1H-NMR spectra acquired in an 800 MHz spectrometer (ChenomxNMRsuite). Their determined concentrations were used for pathway analysis (MetaboAnalyst).