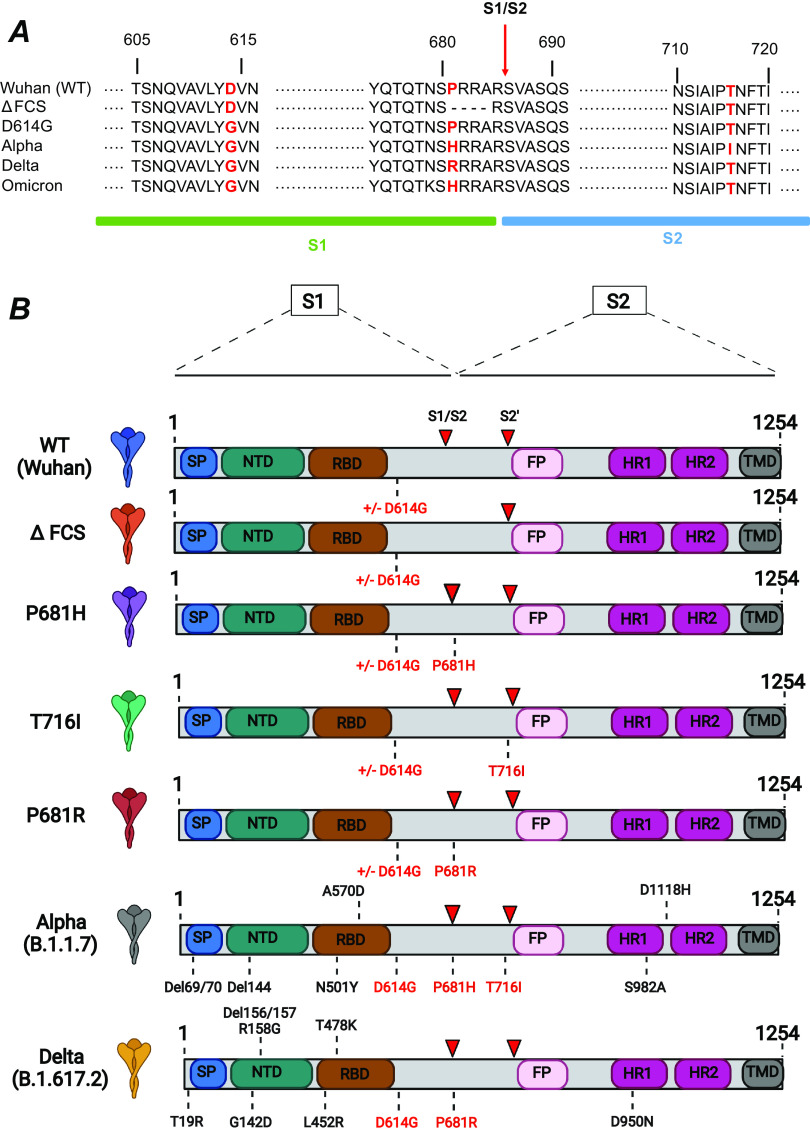
FIG 2.
Spike mutations analyzed in the present study. (A) Sequences surrounding the spike mutations studied. The S1/S2 spike cleavage site is indicated by a vertical arrow. The mutations studied are highlighted in red. The ΔFCS mutation corresponds to the deletion of the PRRA sequence at the S1/S2 cleavage site. (B) Schematic of the spike expression vectors used in the present study. The mutations ΔFCS, P681H, P681R, and T716I were inserted into the Wuhan WT spike backbone truncated at residue 1254, in the presence or absence of the D614G mutation. The spikes containing the full complement of mutations present in the Alpha and Delta VOC were used as controls. SP, signal peptide; NTD, N-terminal domain; RBD, receptor binding domain; FP, fusion peptide; HR, heptad repeat; TMD, transmembrane domain. Red arrowheads indicate the S1/S2 and S2′ cleavage sites.