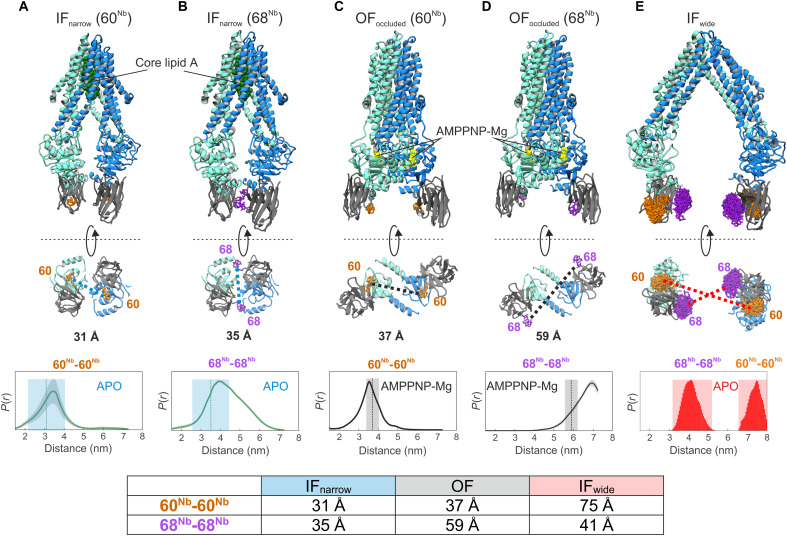
Fig. 1. Structure-based identification and experimental validation of spin-labeling sites.
(A to D) Cryo-EM structures of nanodisc-reconstituted MsbA (cyan/blue) in complex with the nanobody (gray) containing Gd-DOTA either at position 60Nb (A and C) or 68Nb (B and D), obtained in the apo conformation (IFnarrow) or in the presence of AMPPNP-Mg (OFoccluded). Gd-Gd distances are shown as dotted lines in the bottom view of the structures. The bottom of (A) to (D) shows experimentally determined DEER distance distributions using the same samples used for cryo-EM structure determination. Uncertainties estimated from the neural network analysis are shown as gray contour of the distributions (39). Dotted vertical lines denote Gd-Gd distances measured in the respective cryo-EM structures. The blue and gray shaded areas are error margins that correspond to the resolution of the structures and the degree of conformational flexibility determined from multibody refinement as described in Methods. (E) Nanobody docked onto the crystal structure of IFwide MsbA. The in silico predicted rotamers of the Gd-maleimide-DOTA spin label (39) are displayed as orange and purple sticks at nanobody positions 60Nb and 68Nb, respectively. The interspin distances are highlighted with dotted lines. The bottom panel shows simulated distance distributions for the two spin-label pairs of IFwide. Throughout the manuscript, we use the red shaded areas encompassing the full range of simulated distances to schematically represent the distances in the IFwide conformation. A table at the bottom summarizes the mean Gd-Gd distances for the respective conformation.