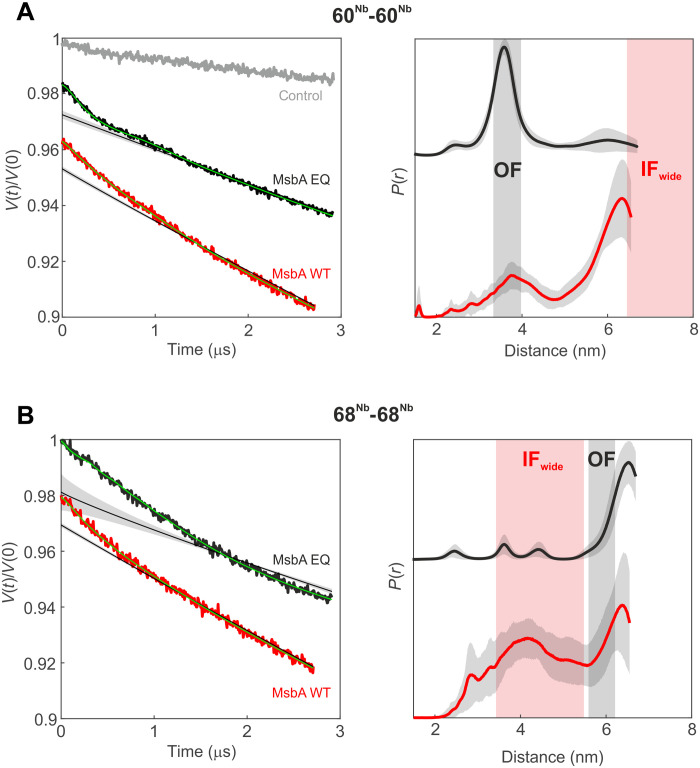
Fig. 4. DEER analysis of MsbA in E. coli cells.
Primary DEER data (left) and corresponding distance distributions (right) of the 60Nb-60Nb pair (A) and the 68Nb-68Nb pair (B) in cells overexpressing mutant (EQ, black) or WT (red) MsbA. A control sample [gray in (A)] with the Gd-labeled nanobody (60Nb) electroporated in cells overexpressing TM287/288 did not show dipolar modulation. The gray shaded areas represent the distance range of the OFoccluded conformation, estimated on the basis of the cryo-EM structures, and the red shaded areas correspond to the distance range predicted for the IFwide conformation using a rotamer approach (see Fig. 1). The obtained distance distributions are further validated by analyses performed with Tikhonov regularization and model-based Gaussian fit (fig. S20).