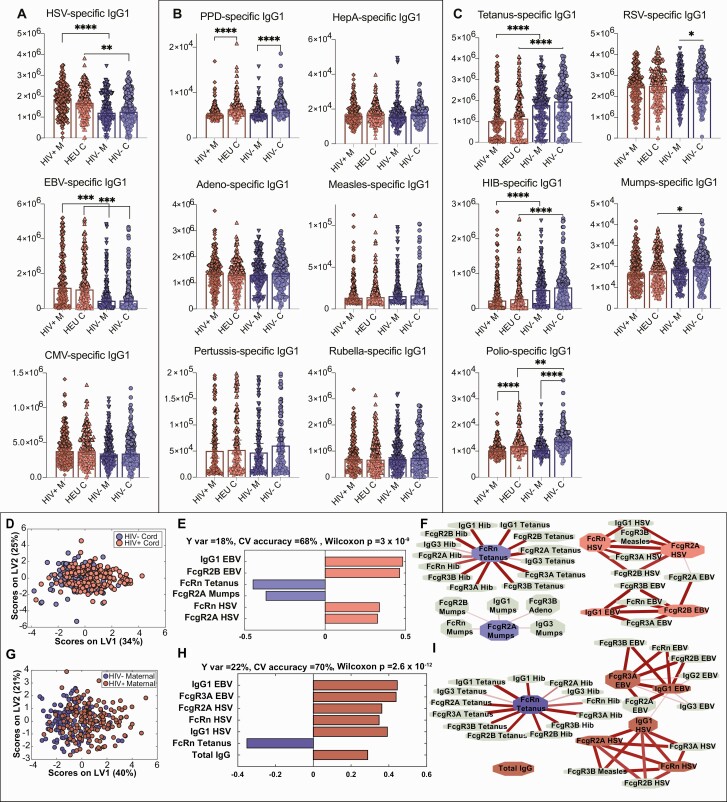
Figure 2.
Differences in human immunodeficiency virus (HIV)–exposed and -unexposed umbilical cord antibody profiles are associated with differences in maternal antibody profiles. A–C, Univariate plots comparing immunoglobulin G1 (IgG1) levels across 4 groups of maternal and umbilical cord samples. HIV+ M indicates maternal plasma from women with HIV (WHIV), HIV– M indicates HIV-uninfected maternal plasma, HEU C indicates HIV-exposed umbilical cord plasma, and HIV– C indicates HIV-unexposed umbilical cord plasma. Only significant comparisons are depicted. Maternal and umbilical cord samples were only compared within serostatus group. To assess significance of the IgG1 level differences between groups, Kruskal-Wallis nonparametric test was applied, followed by Dunn test to correct for multiple comparisons. Adjusted P values: *P < .0332, **P < .0021, ***P < .0002, ****P < .0001. A, Univariate plots showing IgG1 levels for antigen specificities where HIV-uninfected/unexposed maternal and umbilical cord plasma samples are lower than WHIV/HIV-exposed umbilical cord levels for herpesvirus antigens (chronic maternal infections). Herpes simplex virus (HSV)– and Epstein-Barr virus (EBV)–specific IgG1 levels are significantly lower in HIV-uninfected/exposed than WHIV/HIV-exposed, and cytomegalovirus-specific IgG1 differences are not statistically significant. B, Antigen-specific IgG1 levels that are unaffected by maternal HIV serostatus. C, Antigen-specific IgG1 levels that are significantly higher in the HIV-uninfected/unexposed groups than WHIV/HIV-exposed include vaccine-induced antibodies to tetanus, Haemophilus influenzae type b (Hib), and mumps, and nonsignificantly higher for respiratory syncytial virus and polio. D–I, Multivariate analysis to distinguish HIV-exposed and unexposed umbilical cord (D–F) and maternal (G–I) antibody profiles. D, Score plot for the orthogonalized partial least squares discriminant analysis (OPLSDA) analysis using 6 least absolute shrinkage and selection operator (LASSO)–selected features depicts moderately different profiles between HIV-exposed and -unexposed umbilical cord plasma. Latent variable 1 (LV1) captures the separation between HIV-exposed and HIV-unexposed cord plasma (Y var = 18% and X var = 34%), where latent variable 2 (LV2) captures the variance in antibody profiles that does not contribute to this separation (Y var = 0% and X var = 25%). E, Bar graph depicts loadings on LV1 ordered according to their variable importance in projection (VIP) scores and is color-coded to emphasize the features enriched in each group. F, Correlates of the LASSO-selected features are depicted in network format. Only correlation coefficients with multiple comparison corrected P values <.05 and Spearman correlation coefficients >0.3 are included. Line widths and color intensities are proportional to the correlation coefficients. Overall, higher EBV- and HSV-specific antibody titers and associated Fc receptor binding capacity are observed in HIV-exposed umbilical cord plasma, while tetanus-, Hib-, and mumps-specific antibodies are enriched in HIV-unexposed group. G, Similarly, OPLSDA models were built using 7 LASSO-selected antibody features. The dots in the score plot represent WHIV and HIV-uninfected maternal samples. LV1 captures the separation between these maternal groups (Y var = 22% and X var = 40%), and LV2 captures the variance in antibody profiles that does not contribute to this separation (Y var = 0% and X var = 21%). H, Bar graph depicts loadings on LV1 ordered according to their VIP scores and color-coded to emphasize the features enriched in each group. I, Correlates of the LASSO-selected features are depicted in network format with criteria and line depictions similar to (F). Abbreviations: CMV, cytomegalovirus; CV, cross-validation; EBV, Epstein-Barr virus; FcR, Fc receptor; HepA, hepatitis A; HEU, human immunodeficiency virus exposed, uninfected; Hib, Haemophilus influenzae type b; HIV, human immunodeficiency virus; HSV, herpes simplex virus; IgG, immunoglobulin G; LV, latent variable; PPD, purified protein derivative; RSV, respiratory syncytial virus.